Tutorial
Linking NEON aquatic observational and instrument data to answer critical questions in aquatic ecology at the continental scale
Authors: Zachary L. Nickerson
Last Updated: May 16, 2025
Learning Objectives
After completing this activity, you will be able to:
- Download NEON AIS and AOS data using the
neonUtilitiespackage. - Understand downloaded data packages and load them into R for analyses.
- Understand the similarities and linkages between different NEON data products.
- Join data sets within and between data products by standardized variables.
- Plot instrumented and observational data in the same plotting field.
Things You'll Need To Complete This Tutorial
To complete this tutorial you will need R (version >3.4) and, preferably, RStudio loaded on your computer.
Install R Packages
- neonUtilities: Basic functions for accessing NEON data
- neonOS: Basic data wrangling for NEON Observational Data
- tidyverse: Collection of R packages designed for data science
- plotly: Tool for creating interactive, web-based visualizations
- vegan: Functions for analyzing ecological data
- base64enc: Tools for base64 encoding
These packages are on CRAN and can be installed by
install.packages().
Additional Resources
Introduction
Tutorial Overview
This tutorial covers downloading NEON Aquatic Instrument Subsystem (AIS) and
Aquatic Observation Subsystem (AOS) data products using the neonUtilities R
package, as well as basic instruction in beginning to explore and work with the
downloaded data. This includes navigating data packages documentation,
summarizing data for plotting and analysis, combining data within and between
data products, and visualizing AIS and AOS data separately and together.
Helpful Links
Getting started with NEON data: https://www.neonscience.org/resources/getting-started-neon-data-resources
Contact us form: https://www.neonscience.org/about/contact-us
Teaching Modules: https://www.neonscience.org/resources/learning-hub/teaching-modules
QUBES modules: https://qubeshub.org/community/groups/neon/educational_resources
EDDIE modules : https://serc.carleton.edu/eddie/macrosystems/index.html
Spatial data and maps: https://neon.maps.arcgis.com/home/index.html
NEON data portal: https://data.neonscience.org/
NEONScience GitHub repo: https://github.com/NEONScience
SFS 2025 NEON Workshop GitHub repo:
https://github.com/NEONScience/WORKSHOP-SFS-2025
Download Files and Load Directly to R: loadByProduct()
The most popular function in neonUtilities is loadByProduct().
This function downloads data from the NEON API, merges the site-by-month
files, and loads the resulting data tables into the R environment,
assigning each data type to the appropriate R class. This is a popular
choice because it ensures you're always working with the most up-to-date data,
and it ends with ready-to-use tables in R. However, if you use it in
a workflow you run repeatedly, keep in mind it will re-download the
data every time.
Before we get the NEON data, we need to install (if not already done) and load the neonUtilities R package, as well as other packages we will use in the analysis.
# # Install neonUtilities package if you have not yet.
# install.packages("neonUtilities")
# install.packages("neonOS")
# install.packages("tidyverse")
# install.packages("plotly")
# install.packages("vegan")
# install.packages("base64enc")
# Set global option to NOT convert all character variables to factors
options(stringsAsFactors=F)
# Load required packages
library(neonUtilities)
library(neonOS)
library(tidyverse)
library(plotly)
library(vegan)
library(base64enc)
The inputs to loadByProduct() control which data to download and how
to manage the processing. The following are frequently used inputs:
-
dpID: the data product ID, e.g. DP1.20288.001 -
site: defaults to "all", meaning all sites with available data; can be a vector of 4-letter NEON site codes, e.g.c("MART","ARIK","BARC"). -
startdateandenddate: defaults to NA, meaning all dates with available data; or a date in the form YYYY-MM, e.g. 2017-06. Since NEON data are provided in month packages, finer scale querying is not available. Both start and end date are inclusive. -
package: either basic or expanded data package. Expanded data packages generally include additional information about data quality, such as individual quality flag test results. Not every NEON data product has an expanded package; if the expanded package is requested but there isn't one, the basic package will be downloaded. -
release: The data release to be downloaded; either 'current' or the name of a release, e.g. 'RELEASE-2021'. 'current' returns provisional data in addition to the most recent release. To download only provisional data, use release='PROVISIONAL'. Defaults to 'current'. See https://www.neonscience.org/data-samples/data-management/data-revisions-releases for more information. -
include.provisional: Should provisional data be included in the downloaded files? Defaults to F. -
timeIndex: defaults to "all", to download all data; or the number of minutes in the averaging interval. See example below; only applicable to IS data. -
check.size: T or F; should the function pause before downloading data and warn you about the size of your download? Defaults to T; if you are using this function within a script or batch process you will want to set this to F. -
token: this allows you to input your NEON API token to obtain faster downloads. Learn more about NEON API tokens in the Using an API Token when Accessing NEON Data with neonUtilities tutorial.
There are additional inputs you can learn about in the Use the neonUtilities R Package to Access NEON Data tutorial.
The dpID is the data product identifier of the data you want to
download. The DPID can be found on the
Explore Data Products page.
It will be in the form DP#.#####.###. For this tutorial, we'll use some data products collected in NEON's aquatics program:
- DP1.20120.001: Macroinvertebrate collection
- DP4.00130.001: Continuous discharge
Now it's time to consider the NEON field site of interest. If not specified, the default will download a data product from all sites. The following are 4-letter site codes for NEON's 34 aquatics sites as of 2025:
- ARIK = Arikaree River CO
- BARC = Barco Lake FL
- BIGC = Upper Big Creek CA
- BLDE = Black Deer Creek WY
- BLUE = Blue River OK
- BLWA = Black Warrior River AL
- CARI = Caribou Creek AK
- COMO = Como Creek CO
- CRAM = Crampton Lake WI
- CUPE = Rio Cupeyes PR
- FLNT = Flint River GA
- GUIL = Rio Yahuecas PR
- HOPB = Lower Hop Brook MA
- KING = Kings Creek KS
- LECO = LeConte Creek TN
- LEWI = Lewis Run VA
- LIRO = Little Rock Lake WI
- MART = Martha Creek WA
- MAYF = Mayfield Creek AL
- MCDI = McDiffett Creek KS
- MCRA = McRae Creek OR
- OKSR = Oksrukuyik Creek AK
- POSE = Posey Creek VA
- PRIN = Pringle Creek TX
- PRLA = Prairie Lake ND
- PRPO = Prairie Pothole ND
- REDB = Red Butte Creek UT
- SUGG = Suggs Lake FL
- SYCA = Sycamore Creek AZ
- TECR = Teakettle Creek CA
- TOMB = Lower Tombigbee River AL
- TOOK = Toolik Lake AK
- WALK = Walker Branch TN
- WLOU = West St Louis Creek CO
In this exercise, we will pull data from NEON Atlantic Neotropical Domain (D04). The aquatic sites in D04 are Rio Cupeyes (CUPE) and Rio Yahuecas (GUIL). Just substitute the 4-letter site code for any other site at the end of the url.
Now let us download our data. We will focus our exercise on data collected from
2021-10-01 through 2024-09-30 (water years 2022, 2023, 2024). If you are not
using a NEON token to download your data, neonUtilities will ignore the token
input. We set check.size = F so that the script runs well but remember you
always want to check your download size first. For this exercise, we will focus
on the following data products:
AIS Data Products:
- Continuous discharge (DP4.00130.001)
AOS Data Products:
- Macroinvertebrate collection (DP1.20120.001)
Download AOS Data Products
# download data of interest - AOS - Macroinvertebrate collection
inv <- neonUtilities::loadByProduct(dpID="DP1.20120.001",
site=c("CUPE","GUIL"),
startdate="2021-10",
enddate="2024-09",
package="basic",
release= "current",
include.provisional = T,
token = Sys.getenv("NEON_TOKEN"),
check.size = F)
Files Associated with Downloads
The data we've downloaded comes as an object that is a named list of objects.
To work with each of them, select them from the list using the $ operator.
# view all components of the list
names(inv)
## [1] "categoricalCodes_20120" "citation_20120_PROVISIONAL" "citation_20120_RELEASE-2025" "inv_fieldData"
## [5] "inv_persample" "inv_taxonomyProcessed" "issueLog_20120" "readme_20120"
## [9] "validation_20120" "variables_20120"
We can see that there are 10 objects in the downloaded macroinvertebrate collection data.
- Three dataframes of data:
-
inv_fieldData -
inv_persample -
inv_taxonomyProcessed
-
- Five metadata files:
-
categoricalCodes_20120 -
issueLog_20120 -
readme_20120 -
validation_20120 -
variables_20120
-
- Two data citations:
-
citation_20120_PROVISIONAL -
citation_20120_RELEASE-2025
-
If you'd like you can use the $ operator to assign an object from an item in
the list. If you prefer to extract each table from the list and work with it as
independent objects, which we will do, you can use the list2env() function.
# unlist the variables and add to the global environment
list2env(inv,envir = .GlobalEnv)
## <environment: R_GlobalEnv>
Explore: Data Citations
Citing sources correctly helps the NEON user community maintain transparency, openness, and trust, while also providing a benefit of being able to track the impact of NEON on scientific research. Thus, each download of NEON data comes with proper citations custom to to the download that align with NEON's data citation guidelines
# view formatted citations for DP1.20120.001 download
cat(citation_20120_PROVISIONAL)
## @misc{DP1.20120.001/provisional,
## doi = {},
## url = {https://data.neonscience.org/data-products/DP1.20120.001},
## author = {{National Ecological Observatory Network (NEON)}},
## language = {en},
## title = {Macroinvertebrate collection (DP1.20120.001)},
## publisher = {National Ecological Observatory Network (NEON)},
## year = {2025}
## }
cat(`citation_20120_RELEASE-2025`)
## @misc{https://doi.org/10.48443/rmeq-8897,
## doi = {10.48443/RMEQ-8897},
## url = {https://data.neonscience.org/data-products/DP1.20120.001/RELEASE-2025},
## author = {{National Ecological Observatory Network (NEON)}},
## keywords = {diversity, taxonomy, community composition, species composition, population, aquatic, benthic, macroinvertebrates, invertebrates, abundance, streams, lakes, rivers, wadeable streams, material samples, archived samples, biodiversity},
## language = {en},
## title = {Macroinvertebrate collection (DP1.20120.001)},
## publisher = {National Ecological Observatory Network (NEON)},
## year = {2025}
## }
Explore: Metadata
-
categoricalCodes_xxxxx: Some variables in the data tables are published as strings and constrained to a standardized list of values (LOV). This file shows all the LOV options for variables published in this data product.
-
issueLog_xxxxx: Issues that may impact data quality, or changes to a data product that affects all sites, are reported in this file.
-
readme_xxxxx: The readme file provides important information relevant to the data product and the specific instance of downloading the data.
-
validation_xxxxx: If any fields require validation prior to publication, those validation rules are reported in this table
-
variables_xxxxx: This file contains all the variables found in the associated data table(s). This includes full definitions, units, and other important information.
view the entire dataframe in your R environment
view(variables_20120)
Explore: Dataframes
There will always be one or more dataframes that include the primary data of the data product you downloaded. Multiple dataframes are available when there are related datatables for a single data product.
# view the entire dataframe in your R environment
view(inv_fieldData)
Download AIS Data Products
# download data of interest - AIS - Continuous discharge
csd <- neonUtilities::loadByProduct(dpID="DP4.00130.001",
site=c("CUPE","GUIL"),
startdate="2021-10",
enddate="2024-09",
package="basic",
release= "current",
include.provisional = T,
token = Sys.getenv("NEON_TOKEN"),
check.size = F)
Let's see what files are included with an AIS data product download
# view all components of the list
names(csd)
## [1] "categoricalCodes_00130" "citation_00130_PROVISIONAL" "citation_00130_RELEASE-2025" "csd_continuousDischarge"
## [5] "issueLog_00130" "readme_00130" "science_review_flags_00130" "sensor_positions_00130"
## [9] "variables_00130"
This AIS data product contains 1 data table available in the basic package:
-
csd_continuousDischarge- Continuous discharge (streamflow) data at a 1 minute interval. Being a Level 4 data product, this data has been cleaned and gap-filled.
Additionally, there are a couple of metadata file types included in AIS data product downloads that are not included in AOS data product downloads:
- sensor_postions_xxxxx: This file contains information about the coordinates of each sensor, relative to a reference location.
- science_review_flags_xxxxx: This file contains information on quality flags added to the data following expert review for data that are determined to be suspect due to known adverse conditions not captured by automated flagging.
Let's unpack the AIS data product to the environment:
# unlist the variables and add to the global environment
list2env(csd, .GlobalEnv)
## <environment: R_GlobalEnv>
Wrangling AOS Data
The neonOS R package was developed to aid in wrangling NEON Observational
Subsystem (OS) data products. Two functions used in this exercise are:
-
removeDups() -
joinTableNEON()
Removing Duplicates from OS Data
Duplicates can arise in data, but the neonOS::removeDups() function identifies
duplicates in a data table based on primary key information reported in the
variables_xxxxx files included in each data download.
Let's check for duplicates in macroinvertebrate collection data
# what are the primary keys in inv_fieldData?
message("Primary keys in inv_fieldData are: ",
paste(variables_20120$fieldName[
variables_20120$table=="inv_fieldData"
&variables_20120$primaryKey=="Y"
],
collapse = ", ")
)
# identify duplicates in inv_fieldData
inv_fieldData_dups <- neonOS::removeDups(inv_fieldData,
variables_20120)
# what are the primary keys in inv_persample?
message("Primary keys in inv_persample are: ",
paste(variables_20120$fieldName[
variables_20120$table=="inv_persample"
&variables_20120$primaryKey=="Y"
],
collapse = ", ")
)
# identify duplicates in inv_persample
inv_persample_dups <- neonOS::removeDups(inv_persample,
variables_20120)
# what are the primary keys in inv_taxonomyProcessed?
message("Primary keys in inv_taxonomyProcessed are: ",
paste(variables_20120$fieldName[
variables_20120$table=="inv_taxonomyProcessed"
&variables_20120$primaryKey=="Y"
],
collapse = ", ")
)
# identify duplicates in inv_taxonomyProcessed
inv_taxonomyProcessed_dups <- neonOS::removeDups(inv_taxonomyProcessed,
variables_20120)
Thankfully, there are no duplicates in any of the AOS tables used in this exercise!
Joining OS Data Tables
Every NEON data product comes with a Quick Start Guide (QSG). The QSGs contain basic information to help users familiarize themselves with the data products, including description of the data contents, data quality information, common calculations or transformations, and, where relevant, algorithm description and/or table joining instructions.
The QSG for Macroinvertebrate collection can be found on the data product landing page: https://data.neonscience.org/data-products/DP1.20120.001
The neonOS::joinTableNEON() function uses the table joining information in the
QSG to quickly join two related NEON data tables from the same data product
# join inv_fieldData and inv_taxonomyProcessed
inv_fieldTaxJoined <- neonOS::joinTableNEON(inv_fieldData,inv_taxonomyProcessed)
Now, with field and taxonomy data joined. Individual taxon identifications are easily linked to field data such as collection latitude/longitude, habitat type, sampler type, and substratum class.
Wrangling AIS Data
Data from Different Sensor Locations (HOR)
NEON often collects the same type of data from sensors in different locations.
These data are delivered together but you will frequently want to plot the data
separately or only include data from one sensor in your analysis. NEON uses the
horizontalPosition variable in the data tables to describe which sensor
data is collected from. The horizontalPosition is always a three digit number
for AIS data.
The Continuous discharge data product is derived from a single
horizontalPosition, which corresponds to the sensor co-located with the staff
gauge at the site. This is also the location at which all empirical discharge
measurements are taken.
Let's see from which horizontalPosition the Continuous discharge data is
published.
# use dplyr from the tidyverse collection to get all unique horizontal positions
csd_hor <- csd_continuousDischarge%>%
dplyr::distinct(siteID,stationHorizontalID)
print(csd_hor)
## siteID stationHorizontalID
## 1 CUPE 110
## 2 GUIL 110
## 3 GUIL 132
# GUIL has two horizontal positions because the location of the staff gauge
# changed sometime during this time period. At what date did that occur?
max(csd_continuousDischarge$endDate[
csd_continuousDischarge$siteID=="GUIL"
&csd_continuousDischarge$stationHorizontalID=="110"
])
## [1] "2022-12-12 23:58:00 GMT"
At CUPE, the continuous discharge data are published from the 110 position, which is defined as 'water level sensors mounted to a staff gauge at stream sites'.
At GUIL, until 2022-12-12, the continuous discharge data were published from the 110 position. On 2022-12-12, the position changed to 132, which is defined as 'stand-alone water level sensors at downstream (S2) locations at stream sites.'
Average continuous discharge to 15-min interval
To make the continuous discharge data easier to work with for this exercise,
let's use different packages from the tidyverse collection to create a 15-min
averaged table.
# 15-min average of continuous discharge data
CSD_15min <- csd_continuousDischarge%>%
dplyr::mutate(roundDate=lubridate::round_date(endDate,"15 min"))%>%
dplyr::group_by(siteID,roundDate)%>%
dplyr::summarise(dischargeMean=mean(continuousDischarge,na.rm=T),
dischargeCountQF=sum(dischargeFinalQFSciRvw,na.rm = T))
Notice that we included a summation of the science review quality flag (QFSciRvw; binary: 1 = flag, 0 = no flag) fields in the new table.
Plot Data
Now that we have wrangled the data a bit to make it easier to work with, let's make some initial plots to see the AOS and AIS data separately before we begin to investigate questions that involve integrating the data.
AOS Macroinvertebrate abundance and richness
First, we remove the records collected outside of normal sampling bouts as a grab sample. In cases where the NEON field ecologists see interesting organisms that would not be captured using standard field sampling methods, they can collect a grab sample to be identified by the expert taxonomists.
Next, we calculate macroinvertebrate abundance per square meter and taxon richness per sampling bout. This allows us to compare macroinvertebrate data among different samplerTypes and habitatTypes.
We use the vegan R package to calculate richness, evenness, and both the
Shannon and Simpson biodiversity indicies in this exercise. Though we only
focus on richness in the plots, users are encouraged to alter the variables
to view other indices. For a more detailed dive into NEON biodiversity analyses,
see the following NEON tutorial:
Explore and work with NEON biodiversity data from aquatic ecosystems
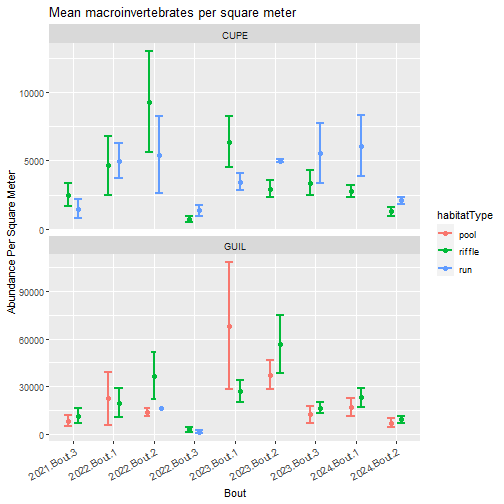
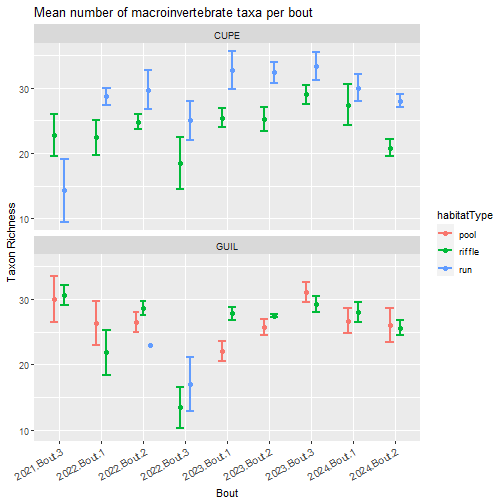
Sampler types (e.g., surber, hand corer, kicknet) are strongly associated with habitat (i.e., riffle, run, pool) and substrata. At some NEON sites, like D04 CUPE, the same sampler is used in two habitat types (surber in riffle and run) because all habitats at the site have the same cobble substrata. Data users should look at the data to determine how they want to discriminate between sampler or habitat type.
For this exercise, we split abundance and richness by habitatType. To split
instead by samplerType, simply do a find+replace of
'habitatType' -> 'samplerType' throughout the code.
### SHOW BREAKDOWN OF SAMPLER TYPE BY HABITAT TYPE AT EACH SITE ###
sampler_habitat_summ <- inv_fieldTaxJoined%>%
dplyr::distinct(siteID,samplerType,habitatType)
sampler_habitat_summ
## siteID samplerType habitatType
## 1 CUPE surber riffle
## 2 CUPE surber run
## 3 GUIL hess pool
## 4 GUIL surber riffle
## 5 GUIL hess run
### PLOT ABUNDANCE ###
# using the `tidyverse` collection, we can clean the data in one piped function
inv_abundance_summ <- inv_fieldTaxJoined%>%
# remove events when no samples were collected (samplingImpractical)
# remove samples not associated with a bout
dplyr::filter(is.na(samplingImpractical)
&!grepl("GRAB|BRYOZOAN",sampleID))%>%
# calculate abundance (individuals per m2^)
dplyr::mutate(abun_M2=estimatedTotalCount/benthicArea)%>%
# clean `collectDate` column header
dplyr::rename(collectDate=collectDate.x)%>%
# first, group including `sampleID` and calculate total abundance per sample
dplyr::group_by(siteID,collectDate,eventID,sampleID,habitatType,boutNumber)%>%
dplyr::summarize(abun_M2_sum = sum(abun_M2, na.rm = TRUE))%>%
# second, group excluding `sampleID` to summarize by each bout (`eventID`)
dplyr::group_by(siteID,collectDate,eventID,habitatType,boutNumber)%>%
# summarize to get mean (+/- se) abundance by bout and sampler type
dplyr::summarise_each(funs(mean,sd,se=sd(.)/sqrt(n())))%>%
# get categorical variable to sort bouts chronologically
dplyr::mutate(year=substr(eventID, 6,9),
yearBout=paste(year,"Bout",boutNumber, sep = "."))
# produce stacked plot to show trends within and across sites
inv_abundance_summ%>%
ggplot2::ggplot(aes(fill=habitatType, color=habitatType, y=abun_M2_sum_mean, x=yearBout))+
ggplot2::geom_point(position=position_dodge(0.5), size=2)+
ggplot2::geom_errorbar(aes(ymin=abun_M2_sum_mean-abun_M2_sum_se,
ymax=abun_M2_sum_mean+abun_M2_sum_se),
width=0.4, alpha=3.0, linewidth=1,
position = position_dodge(0.5))+
ggplot2::facet_wrap(~siteID,ncol = 1,scales="free_y")+
ggplot2::theme(axis.text.x = element_text(size = 10, angle = 30,
hjust = 1, vjust = 1))+
ggplot2::labs(title = "Mean macroinvertebrates per square meter",
y = "Abundance Per Square Meter",
x = "Bout")
### PLOT RICHNESS ###
inv_richness_clean <- inv_fieldTaxJoined%>%
# remove events when no samples were collected (samplingImpractical)
# remove samples not associated with a bout
dplyr::filter(is.na(samplingImpractical)
&!grepl("GRAB|BRYOZOAN",sampleID))%>%
# clean `collectDate` column header
dplyr::rename(collectDate=collectDate.x)
# extract sample metadata
inv_sample_info <- inv_richness_clean%>%
dplyr::select(sampleID, domainID, siteID, namedLocation,
collectDate, eventID, boutNumber,
habitatType, samplerType, benthicArea)%>%
dplyr::distinct()
# filter out rare taxa: only observed 1 (singleton) or 2 (doubleton) times
inv_rare_taxa <- inv_richness_clean%>%
dplyr::distinct(sampleID, acceptedTaxonID, scientificName)%>%
dplyr::group_by(scientificName)%>%
dplyr::summarize(occurrences = n())%>%
dplyr::filter(occurrences > 2)
# filter richness table based on taxon list excluding singletons and doubletons
inv_richness_clean <- inv_richness_clean %>%
dplyr::filter(scientificName%in%inv_rare_taxa$scientificName)
# create a matrix of taxa by sampleID
inv_richness_clean_wide <- inv_richness_clean %>%
# subset to unique combinations of `sampleID` and `scientificName`
dplyr::distinct(sampleID,scientificName,.keep_all = T)%>%
# remove any records with no abundance data
dplyr::mutate(abun_M2=estimatedTotalCount/benthicArea)%>%
filter(!is.na(abun_M2))%>%
# pivot to wide format, sum multiple counts per sampleID
tidyr::pivot_wider(id_cols = sampleID,
names_from = scientificName,
values_from = abun_M2,
values_fill = list(abun_M2 = 0),
values_fn = list(abun_M2 = sum)) %>%
tibble::column_to_rownames(var = "sampleID") %>%
#round to integer so that vegan functions will run
round()
# code check - check col and row sums
# mins should all be > 0 for further analysis in vegan
if(colSums(inv_richness_clean_wide) %>% min()==0){
stop("Column sum is 0: do not proceed with richness analysis!")
}
if(rowSums(inv_richness_clean_wide) %>% min()==0){
stop("Row sum is 0: do not proceed with richness analysis!")
}
# use the `vegan` package to calculate diversity indices
# calculate richness
inv_richness <- as.data.frame(
vegan::specnumber(inv_richness_clean_wide)
)
names(inv_richness) <- "richness"
inv_richness_stats <- vegan::estimateR(inv_richness_clean_wide)
# calculate evenness
inv_evenness <- as.data.frame(
vegan::diversity(inv_richness_clean_wide)/
log(vegan::specnumber(inv_richness_clean_wide))
)
names(inv_evenness) <- "evenness"
# calculate shannon index
inv_shannon <- as.data.frame(
vegan::diversity(inv_richness_clean_wide, index = "shannon")
)
names(inv_shannon) <- "shannon"
# calculate simpson index
inv_simpson <- as.data.frame(
vegan::diversity(inv_richness_clean_wide, index = "simpson")
)
names(inv_simpson) <- "simpson"
# create a single data frame
inv_diversity_indices <- cbind(inv_richness, inv_evenness, inv_shannon, inv_simpson)
# bring in the metadata table created earlier
inv_diversity_indices <- dplyr::left_join(
tibble::rownames_to_column(inv_diversity_indices),
inv_sample_info,
by = c("rowname" = "sampleID")) %>%
dplyr::rename(sampleID = rowname)
# create summary table for plotting
inv_diversity_summ <- inv_diversity_indices%>%
tidyr::pivot_longer(c(richness,evenness,shannon,simpson),
names_to = "indexName",
values_to = "indexValue")%>%
group_by(siteID,collectDate,eventID,habitatType,boutNumber,indexName)%>%
dplyr::summarize(mean = mean(indexValue),
n=n(),
sd = sd(indexValue),
se=sd/sqrt(n))%>%
dplyr::mutate(year=substr(eventID, 6,9),
yearBout=paste(year,"Bout",boutNumber, sep = "."))
# produce plot to show trends within and across sites
inv_diversity_summ%>%
dplyr::filter(indexName=="richness")%>%
ggplot2::ggplot(aes(fill=habitatType, color=habitatType, y=mean, x=yearBout))+
ggplot2::geom_point(position=position_dodge(0.5), size=2)+
ggplot2::geom_errorbar(aes(ymin=mean-se, ymax=mean+se),
width=0.4, alpha=3.0, linewidth=1,
position = position_dodge(0.5))+
ggplot2::facet_wrap(~siteID,ncol=1)+
ggplot2::theme(axis.text.x = element_text(size = 10, angle = 30,
hjust = 1, vjust = 1))+
labs(title="Mean number of macroinvertebrate taxa per bout",
y= "Taxon Richness", x = "Bout")
AIS Continuous discharge timseries
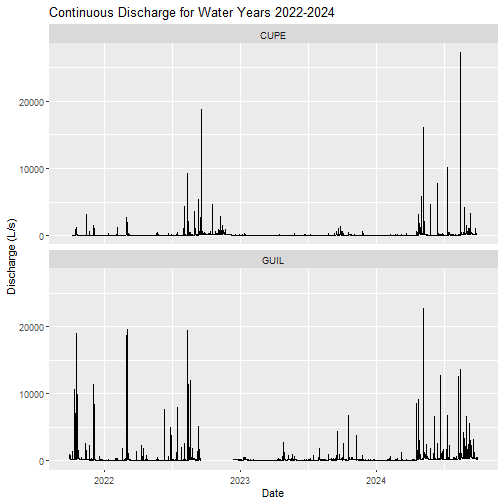
Now, let's visualize the cleaned and gap-filled continuous discharge timeseries for the two NEON D04 sites.
CSD_15min%>%
ggplot2::ggplot(aes(x=roundDate,y=dischargeMean))+
ggplot2::geom_line()+
ggplot2::facet_wrap(~siteID,ncol = 1)+
# ggplot2::scale_y_log10()+ # Include to show discharge axis in log scale
labs(title="Continuous Discharge for Water Years 2022-2024",
y= "Discharge (L/s)", x = "Date")
Visualize AOS and AIS Data Together
Next, we will use the R package plotly to make fun interactive plots allowing
us to view AOS and AIS data in the same plotting field. There is a
lot of code here to correctly format the plot in a way to provide as much info
and be as interactive as possible in a single plotting field.
The plotly package allows us to interact with the plots in the following ways:
- Zoom and pan along the x- and y-axes
- Switch discharge timeseries between linear and log scale
- Turn on/off traces in the plot by clicking the legend entries
- Scroll long each individual y-axis
INV Abundance and Richness + Discharge Over Time
Click on traces to display or hide them. (Note: INV traces defaulted to hidden)
# choose the site(s) you want to plot
siteToPlot <- c("CUPE","GUIL")
for(s in 1:length(siteToPlot)){
# begin the plot code
AOS_AIS_plot <- CSD_15min%>%
dplyr::filter(siteID==siteToPlot[s])%>%
plotly::plot_ly()%>%
# add trace for continuous discharge
plotly::add_trace(x=~roundDate,y=~dischargeMean,
type="scatter",mode="line",
line=list(color = 'darkgray'),
name="Discharge")%>%
# add trace for INV abundance
plotly::add_trace(data=inv_abundance_summ%>%
dplyr::filter(siteID==siteToPlot[s]),
x=~collectDate,y=~abun_M2_sum_mean,
split=~paste0("INV Abundance: ",habitatType),
yaxis="y2",type="scatter",mode="line",
error_y=~list(array=abun_M2_sum_se,
color='darkorange'),
marker=list(color="darkorange"),
line=list(color="darkorange"),
visible="legendonly")%>%
# add trace for INV richness
plotly::add_trace(data=inv_diversity_summ%>%
dplyr::filter(siteID==siteToPlot[s]
&indexName=="richness"),
x=~collectDate,y=~mean,
split=~paste0("INV Richness: ",habitatType),
yaxis="y3",type="scatter",mode="line",
error_y=~list(array=se,
color='darkgreen'),
marker=list(color="darkgreen"),
line=list(color="darkgreen"),
visible="legendonly")%>%
# define the layout of the plot
plotly::layout(
title = paste0(siteToPlot[s],
" Discharge w/ Macroinvertebrate Abundance & Richness"),
# format x-axis
xaxis=list(title="dateTime",
automargin=TRUE,
domain=c(0,0.9)),
# format first y-axis
yaxis=list(
side='left',
title='Discharge (L/s)',
showgrid=FALSE,
zeroline=FALSE,
automargin=TRUE),
# format second y-axis
yaxis2=list(
side='right',
overlaying="y",
title='INV Abundance',
showgrid=FALSE,
automargin=TRUE,
zeroline=FALSE,
tickfont=list(color = 'darkorange'),
titlefont=list(color = 'darkorange')),
# format third y-axis
yaxis3=list(
side='right',
overlaying="y",
anchor="free",
title='INV Richness',
showgrid=FALSE,
zeroline=FALSE,
automargin=TRUE,
tickfont=list(color = 'darkgreen'),
titlefont=list(color = 'darkgreen'),
position=0.99),
# format legend
legend=list(xanchor = 'center',
yanchor = 'top',
orientation = 'h',
x=0.5,y=-0.2),
# add button to switch discharge between linear and log
updatemenus=list(
list(
type='buttons',
buttons=list(
list(label='linear',
method='relayout',
args=list(list(yaxis=list(type='linear')))),
list(label='log',
method='relayout',
args=list(list(yaxis=list(type='log'))))))))
assign(paste0("AOS_AIS_plot_",siteToPlot[s]),AOS_AIS_plot)
}
# show plot at CUPE
AOS_AIS_plot_CUPE

# show plot at GUIL
AOS_AIS_plot_GUIL

What kind of observations can be made when examining AIS discharge and AOS macroinvertebrate data on the same plotting field at NEON's two neotropical aquatic sites?
The standardized spatiotemporal design of NEON's aquatic data products allows one to easily run the same analysis for any NEON site. Given that all of NEON's 24 stream sites publish both AIS continuous discharge and AOS macroinvertebrate collection data products, users can substitute any two NEON stream site IDs into this exercise to assess the relationship between stream discharge and macroinvertebrate abundance and richness.
Visit the Explore NEON Field Sites webpage to learn more about the different NEON aquatic sites. To run this exercise on a different combination of two sites, use find+replace to change the site IDs throughout the code.
Further Exploration
Now, we can take what we have learned about NEON AOS and AIS data and look at other case studies using different NEON data products.
Case Study 1: Examine relationships between discharge, sediment particle size distribution, and macroinvertebrate abundance/diversity at 2 sites impacted by hurricanes.
In September 2022, Hurricane Fiona struck land in Puerto Rico as a Category 1 hurricane. Two NEON D04 aquatic sites were impacted. Here, we scale three data products across time to get an integrated look at how Hurricane Fiona (red line) affected the hydrology, morphology, and biology of two streams.
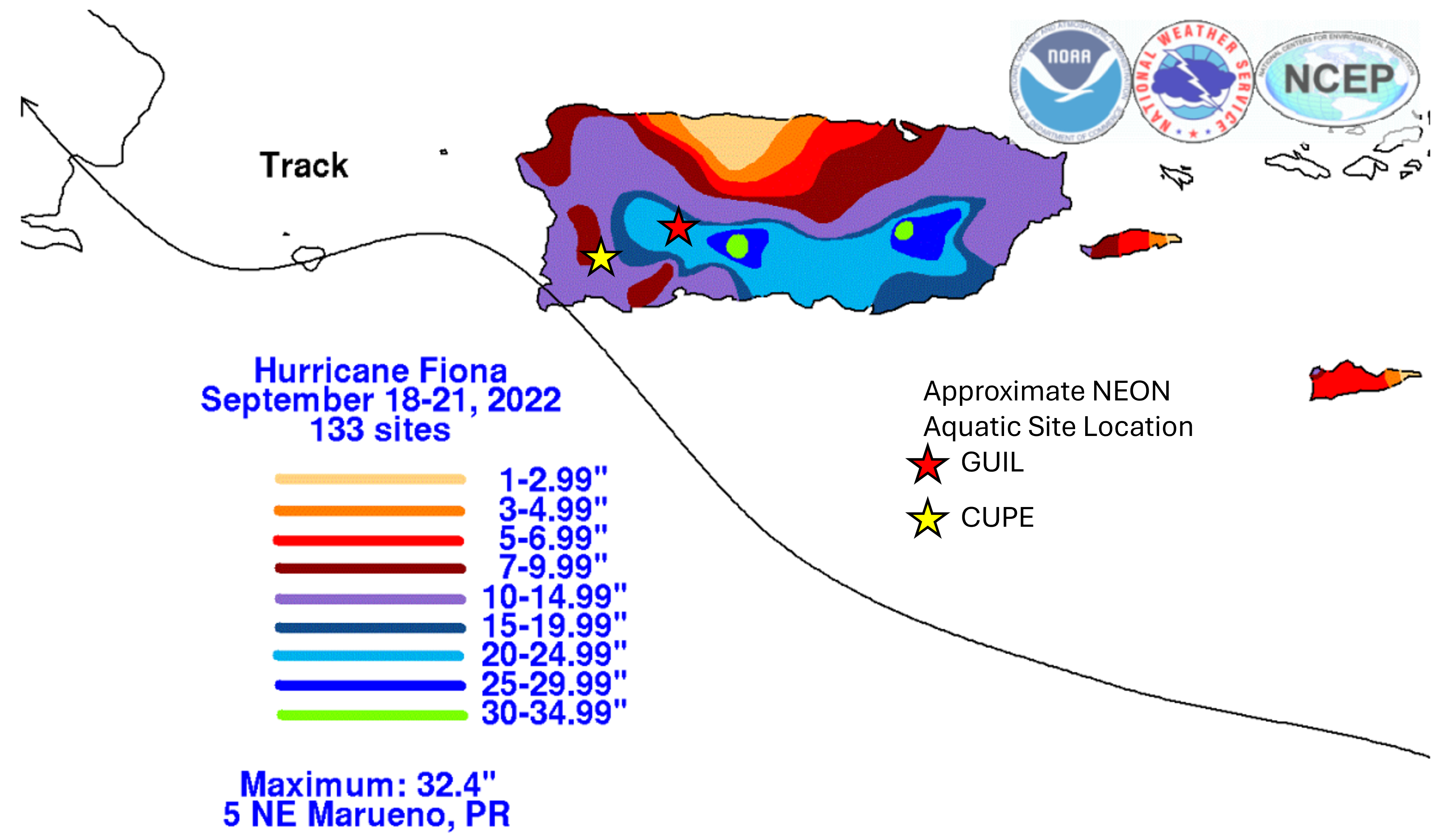
Total rainfall accumulation in Puerto Rico from Hurricane Fiona, overlaid with approximate locations of the two NEON D04 aquatic sites: CUPE, GUIL. Image source: https://www.nhc.noaa.gov/data/tcr/AL072022_Fiona.pdf
For this case study, we will look again at the relationship between stream discharge and macroinvertebrate abundance and richness with the hurricane event highlighted. We will compare the effects of the hurricane on stream hydrology and biology between the two sites.
We will also bring in a third NEON aquatic data product:
- Stream morphology maps (DP4.00131.001)
From the Stream morphology maps data product, we will examine the effect of Hurricane Fiona on streambed particle size distribution, expanding our exploration of NEON aquatic data to uncover linkages between the hydrology, morphology, and biology of NEON streams.
According to NOAA, Hurricane Fiona traveled through Puerto Rico between 18-21 September, 2022. Let's highlight that event in our combined AOS and AIS plots by adding a red vertical dashed line on 2022-09-19.
# identify the date of Fiona
fionaDate <- "2022-09-19"
# highlight Fiona at CUPE
AOS_AIS_plot_CUPE_Fiona <- AOS_AIS_plot_CUPE%>%
# add dashed vertical line to plot created in previous exercise
plotly::add_segments(x=as.POSIXct(fionaDate,tz="UTC"),
xend=as.POSIXct(fionaDate,tz="UTC"),
y=0,
yend=max(CSD_15min$dischargeMean[
CSD_15min$siteID=="CUPE"],
na.rm = T),
name="Fiona",
line=list(color='red',dash='dash'))
# highlight Fiona at GUIL
AOS_AIS_plot_GUIL_Fiona <- AOS_AIS_plot_GUIL%>%
# add dashed vertical line to plot created in previous exercise
plotly::add_segments(x=as.POSIXct(fionaDate,tz="UTC"),
xend=as.POSIXct(fionaDate,tz="UTC"),
y=0,
yend=max(CSD_15min$dischargeMean[
CSD_15min$siteID=="CUPE"],
na.rm = T),
name="Fiona",
line=list(color='red',dash='dash'))
Next, we will use the neonUtilities function loadByProduct() to load data
from the Stream morphology maps data product into R.
# download data of interest - AOS - Stream morphology maps
# the expanded download package is needed to read in the geo_pebbleCount table
geo <- neonUtilities::loadByProduct(dpID="DP4.00131.001",
site=c("CUPE","GUIL"),
startdate="2021-10",
enddate="2024-09",
package="expanded",
release= "current",
include.provisional = T,
token = Sys.getenv("NEON_TOKEN"),
check.size = F)
# unlist the variables and add to the global environment
list2env(geo,envir = .GlobalEnv)
There are many data tables included in this Level 4 AOS download package, but we are only interested in using one table for this exercise:
-
geo_pebbleCount- Sediment particle size data collected during pebble count sampling. Pebble count surveys are conducted once per year, typically during periods of baseflow at a given site.
Check for duplicates in geo_pebbleCount using neonOS::removeDups().
# what are the primary keys in geo_pebbleCount?
message("Primary keys in geo_pebbleCount are: ",
paste(variables_00131$fieldName[
variables_00131$table=="geo_pebbleCount"
&variables_00131$primaryKey=="Y"
],
collapse = ", ")
)
# identify duplicates in geo_pebbleCount
geo_pebbleCount_dups <- neonOS::removeDups(geo_pebbleCount,
variables_00131)
There are no duplicates! Let's proceed.
Next, let's wrangle the data and plot cumulative frequency curves to visualize particle size distributions for NEON D04 aquatic sites across water years 2022, 2023, and 2024.
# we want to plot the frequency of `pebbleSize`
# `pebbleSize` is published as a categorical variable (range of size - mm)
# For plotting purposes, convert `pebbleSize` to numeric (lower number in range)
geo_pebbleCount$pebbleSize_num <- NA
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="< 2 mm: silt/clay"
] <- 0
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="< 2 mm: sand"
] <- 0
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="2 - 2.8 mm: very coarse sand"
] <- 2
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="2.8 - 4 mm: very fine gravel"
] <-2.8
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="4 - 5.6 mm: fine gravel"
] <- 4
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="5.6 - 8 mm: fine gravel"
] <- 5.6
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="8 - 11 mm: medium gravel"
] <- 8
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="11 - 16 mm: medium gravel"
] <- 11
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="16 - 22.6 mm: coarse gravel"
] <- 16
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="22.6 - 32 mm: coarse gravel"
] <- 22.6
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="32 - 45 mm: very coarse gravel"
] <- 32
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="45 - 64 mm: very coarse gravel"
] <- 45
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="64 - 90 mm: small cobble"
] <- 64
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="90 - 128 mm: medium cobble"
] <- 90
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="128 - 180 mm: large cobble"
] <- 128
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="180 - 256 mm: large cobble"
] <- 180
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="> 256 mm: boulder"
] <- 256
geo_pebbleCount$pebbleSize_num[
geo_pebbleCount$pebbleSize=="> 256 mm: bedrock"
] <- 256
# each 'siteID' and 'surveyEndDate' represents a unique pebble count survey
# group by 'siteID' and 'surveyEndDate'` and calculate frequency
geo_pebbleCount_freq <- geo_pebbleCount%>%
dplyr::group_by(siteID,surveyEndDate,eventID,pebbleSize_num)%>%
dplyr::summarise(frequency=n()/200)
# calculate a cumulative sum of frequency per event ID
for(e in 1:length(unique(geo_pebbleCount_freq$eventID))){
eventID_freq <- geo_pebbleCount_freq%>%
filter(eventID==unique(geo_pebbleCount$eventID)[e])
eventID_freq$CumulativeFreq <- cumsum(eventID_freq$frequency)*100
if(e==1){
geo_pebbleCount_freqCumm <- eventID_freq
}else{
geo_pebbleCount_freqCumm <- rbind(geo_pebbleCount_freqCumm,eventID_freq)
}
}
# assign a year to each survey
geo_pebbleCount_freqCumm <- geo_pebbleCount_freqCumm%>%
dplyr::mutate(year=format(surveyEndDate,"%Y"))
# create cumulative frequency curve plot using `geom_smooth`
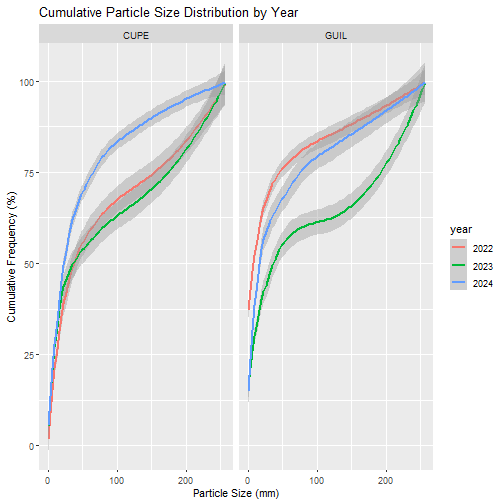
geo_pebbleCount_freqCumm%>%
ggplot2::ggplot(aes(x = pebbleSize_num, y = CumulativeFreq, color = year)) +
ggplot2::geom_smooth(method = "loess", se = T, linewidth = 0.75) +
ggplot2::labs(title="Cumulative Particle Size Distribution by Year",
x = "Particle Size (mm)", y = "Cumulative Frequency (%)") +
ggplot2::facet_wrap(~siteID)
To effectively view the particle size distribution data with the other two data
products, we will embed them as ggplot subplots in the larger plotly plot.
# generate small, simple subplots of each pebble count survey
# loop through each site and year to make plot and save to the working directory
# for(s in 1:length(unique(geo_pebbleCount_freqCumm$siteID))){
# currSite <- unique(geo_pebbleCount_freqCumm$siteID)[s]
# for(y in 1:length(unique(geo_pebbleCount_freqCumm$year))){
# currYear <- unique(geo_pebbleCount_freqCumm$year)[y]
# currPlot <- geo_pebbleCount_freqCumm%>%
# dplyr::filter(siteID==currSite
# &year==currYear)%>%
# ggplot2::ggplot(aes(x = pebbleSize_num, y = CumulativeFreq)) +
# ggplot2::geom_smooth(method = "loess", se = T, linewidth = 0.75) +
# ggplot2::labs(x = NULL, y = NULL)+
# ggplot2::scale_y_continuous(limits=c(0,105))+
# ggplot2::scale_x_continuous(limits=c(0,260))+
# ggplot2::theme_classic()+
# ggplot2::theme(text=element_text(size=18))
# ggplot2::ggsave(plot=currPlot,
# filename=paste0("images/psd_",currSite,currYear,".png"),
# width = 4, height = 7, units = "cm")
# }
# }
# re-generate the CUPE plot with particle size distribution subplots added
AOS_AIS_plot_CUPE_Fiona%>%
layout(images=list(
# show the CUPE 2022 pebble count survey, conducted 2022-04
list(source=base64enc::dataURI(file="images/psd_CUPE2022.png"),
x = 0.05, y = 0.7,
sizex = 0.25, sizey = 0.25,
xref = "paper", yref = "paper",
xanchor = "left", yanchor = "bottom"),
# show the CUPE 2023 pebble count survey, conducted 2022-05
list(source=base64enc::dataURI(file="images/psd_CUPE2023.png"),
x = 0.4, y = 0.7,
sizex = 0.25, sizey = 0.25,
xref = "paper", yref = "paper",
xanchor = "left", yanchor = "bottom"),
# show the CUPE 2024 pebble count survey, conducted 2022-05
list(source=base64enc::dataURI(file="images/psd_CUPE2024.png"),
x = 0.7, y = 0.7,
sizex = 0.25, sizey = 0.25,
xref = "paper", yref = "paper",
xanchor = "left", yanchor = "bottom")
))

# re-generate the GUIL plot with particle size distribution subplots added
AOS_AIS_plot_GUIL_Fiona%>%
layout(images=list(
# show the GUIL 2022 pebble count survey, conducted 2022-04
list(source=base64enc::dataURI(file="images/psd_GUIL2022.png"),
x = 0.05, y = 0.7,
sizex = 0.25, sizey = 0.25,
xref = "paper", yref = "paper",
xanchor = "left", yanchor = "bottom"),
# show the GUIL 2023 pebble count survey, conducted 2023-03
list(source=base64enc::dataURI(file="images/psd_GUIL2023.png"),
x = 0.35, y = 0.7,
sizex = 0.25, sizey = 0.25,
xref = "paper", yref = "paper",
xanchor = "left", yanchor = "bottom"),
# show the GUIL 2024 pebble count survey, conducted 2024-07
list(source=base64enc::dataURI(file="images/psd_GUIL2024.png"),
x = 0.75, y = 0.7,
sizex = 0.25, sizey = 0.25,
xref = "paper", yref = "paper",
xanchor = "left", yanchor = "bottom")
))

Discussion: With the three data products viewed together in relation to the Hurricane Fiona event, there are several observations that can be made:
-
Hydrology
- Both sites were hit with heavy rain, with CUPE discharge reaching nearly 20,000 L/s at its peak.
- Sensor infrastructure at GUIL was temporarily compromised during the storm, resulting in a data gap.
-
Morphology
- The sediment particle size distribution data suggests that CUPE sediment remained relatively stable pre- and post-storm, while GUIL showed heavier loss of small sediment via scouring.
-
Biology
- While both sites show a large decrease in macroinvertebrate abundance post-storm, GUIL shows a more negative effect to macroinvertebrate diversity relative to CUPE.
- Both sites show a trend back to pre-storm abundance and diversity numbers by the following spring bout.
By integrating data products across time, we observe disparate effects of Hurricane Fiona on NEON D04 sites, with a potential relationship being revealed between the stability of the streambed substrate and the loss of macroinvertebrate diversity immediately following a major precipitation event.
Case Study 2: Can relationships between sensor measurements and discrete water chemistry data be used to expand the temporal extent of ecologically-relevant analytes?
We are going to switch gears to a different kind of integration between AOS and AIS data. We evaluate the relationship between high-frequency fluorescent dissolved organic matter (fDOM) AIS data and dissolved organic carbon (DOC) data analyzed from AOS water chemistry grab samples to model a continuous DOC timeseries at D04 CUPE.
The data products downloaded here are:
AIS Data Products:
- Water quality (DP1.20288.001)
AOS Data Products:
- Chemical properties of surface water (DP1.20093.001)
First, download AOS data, run the duplicate check, and plot the data. We will stick with the 2021-10-01 to 2024-09-30 time range.
# download data of interest - AOS - Chemical properties of surface water
swc <- neonUtilities::loadByProduct(dpID="DP1.20093.001",
site=c("CUPE"),
startdate="2021-10",
enddate="2024-09",
package="basic",
release= "current",
include.provisional = T,
token = Sys.getenv("NEON_TOKEN"),
check.size = F)
# unlist the variables and add to the global environment
list2env(swc,envir = .GlobalEnv)
The data table we are interested in here is swc_externalLabDataByAnalyte:
"Long-format results of chemical analysis of up to 28 unique analytes from
surface water and groundwater grab samples."
# check if there are duplicate DOC records
# what are the primary keys in swc_externalLabDataByAnalyte?
message("Primary keys in swc_externalLabDataByAnalyte are: ",
paste(variables_20093$fieldName[
variables_20093$table=="swc_externalLabDataByAnalyte"
&variables_20093$primaryKey=="Y"
],
collapse = ", ")
)
# identify duplicates in swc_externalLabDataByAnalyte
swc_externalLabDataByAnalyte_dups <- neonOS::removeDups(
swc_externalLabDataByAnalyte,
variables_20093)
# no duplicates, great!
# lab data is published `long-format` with 28 analytes analyzed
# show all the analytes published in the lab data
print(unique(swc_externalLabDataByAnalyte$analyte))
## [1] "TDP" "SO4" "TP" "NH4 - N" "Mg"
## [6] "NO2 - N" "F" "Si" "TDS" "UV Absorbance (254 nm)"
## [11] "Cl" "Ca" "TN" "UV Absorbance (280 nm)" "NO3+NO2 - N"
## [16] "TSS" "TPC" "TDN" "Na" "Br"
## [21] "Mn" "Fe" "DOC" "DIC" "K"
## [26] "Ortho - P" "TOC" "TPN"
# for this exercise, subset lab data to only dissolved organic carbon (DOC)
DOC <- swc_externalLabDataByAnalyte%>%
dplyr::filter(analyte=="DOC")
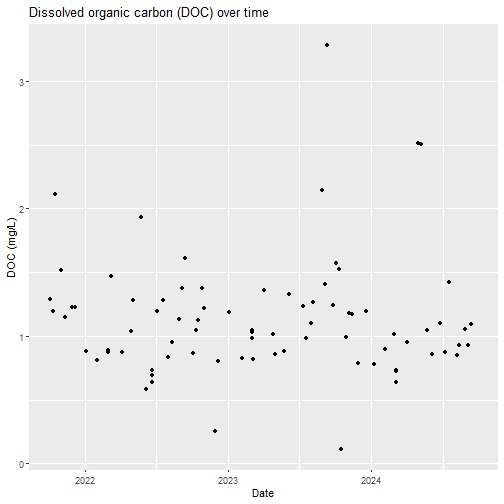
# plot a timeseries of DOC
DOC_plot <- DOC%>%
ggplot2::ggplot(aes(x=collectDate,y=analyteConcentration))+
ggplot2::geom_point()+
ggplot2::labs(title = "Dissolved organic carbon (DOC) over time",
y = "DOC (mg/L)",
x = "Date")
DOC_plot
Next, download AIS data, subset to the appropriate horizontalPosition, wrangle
the data for analysis, and plot the data.
# download data of interest - AIS - Water quality
waq <- neonUtilities::loadByProduct(dpID="DP1.20288.001",
site=c("CUPE"),
startdate="2021-10",
enddate="2024-09",
package="basic",
release= "current",
include.provisional = T,
token = Sys.getenv("NEON_TOKEN"),
check.size = F)
# unlist the variables and add to the global environment
list2env(waq,envir = .GlobalEnv)
The data table we are interested in here is waq_instantaneous: "Wide-format table
published many water quality metrics in wide-format, including fDOM, dissolved oxygen,
specific conductance, pH, chlorophyll, and turbidity."
# `waq_instantaneous` table published many water quality metrics in wide-format
# other than fDOM, many other metrics are published in `waq_instantaneous`
# including: dissolved oxygen, specific conductance, pH, chlorophyll, turbidity
# according to the NEON AIS spatial design, fDOM is only measured at the
# downstream sensor set (S2, HOR = 102); subset to HOR 102
WAQ_102 <- waq_instantaneous%>%
dplyr::filter(horizontalPosition==102)
# `waq_instantaneous` is published at a 1 minute temporal resolution
# for ease of plotting, let's create a 15-minute average table
fDOM_15min <- WAQ_102%>%
# remove NULL records
dplyr::filter(!is.na(rawCalibratedfDOM))%>%
# remove records with a final QF
dplyr::filter(fDOMFinalQF==0)%>%
# create 15-minute average of fDOM
mutate(roundDate=lubridate::round_date(endDateTime,"15 min"))%>%
group_by(siteID,roundDate)%>%
summarize(mean_fDOM=mean(rawCalibratedfDOM))
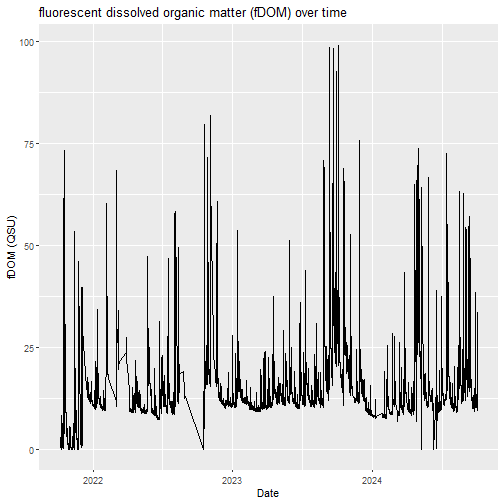
# plot a timeseries of fDOM
fDOM_plot <- fDOM_15min%>%
ggplot2::ggplot(aes(x=roundDate,y=mean_fDOM))+
ggplot2::geom_line()+
ggplot2::labs(title = "fluorescent dissolved organic matter (fDOM) over time",
y = "fDOM (QSU)",
x = "Date")
fDOM_plot
Both data products are published in Coordinated Universal Time (UTC), as are all AOS and AIS data, which makes joining across tables easy. Let's join the AOS and AIS data into a single data frame from which we will model the two variables.
# round DOC `collectDate` to the nearest 15 minute timestamp
DOC$roundDate <- lubridate::round_date(DOC$collectDate,"15 min")
# perform a left-join, which will join an AIS DOC record to every AIS fDOM
# record based on matching timestamps
fDOM_DOC_join <- dplyr::left_join(fDOM_15min,DOC,by="roundDate")
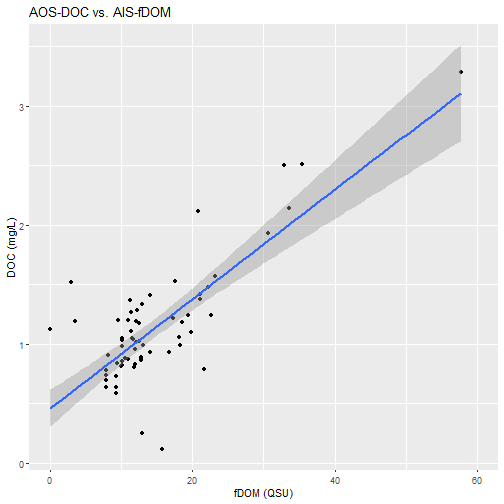
Create a linear regression to analyze the correlation of the two variables
# use `lm` function to create a linear regression: DOC~fDOM
model <- lm(analyteConcentration~mean_fDOM,data=fDOM_DOC_join)
# view a summary of the regression model
print(summary(model))
##
## Call:
## lm(formula = analyteConcentration ~ mean_fDOM, data = fDOM_DOC_join)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.0706 -0.1706 -0.0468 0.1403 0.9160
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.459889 0.078252 5.877 1.64e-07 ***
## mean_fDOM 0.045863 0.004593 9.986 1.12e-14 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.3208 on 64 degrees of freedom
## (81058 observations deleted due to missingness)
## Multiple R-squared: 0.6091, Adjusted R-squared: 0.603
## F-statistic: 99.71 on 1 and 64 DF, p-value: 1.117e-14
# show a plot of the relationship with a linear trendline added
fDOM_DOC_plot <- fDOM_DOC_join%>%
ggplot2::ggplot(aes(x=mean_fDOM,y=analyteConcentration))+
ggplot2::geom_point()+
ggplot2::geom_smooth(method="lm",se=T)+
ggplot2::scale_x_continuous(limits=c(0,60))+
ggplot2::labs(title = "AOS-DOC vs. AIS-fDOM",
y = "DOC (mg/L)",
x = "fDOM (QSU)")
fDOM_DOC_plot
Given relatively high AIS data completeness and a correlative relationship between AOS-DOC and AIS-fDOM, Let's model DOC vs. fDOM from 2021-10-01 to 2024-09-30. We will add modelled continuous DOC as a column in the joined table.
# predict continuous doc based on the linear regression model coefficients
fDOM_DOC_join$fit <- predict(model,
newdata = fDOM_DOC_join,
interval = "confidence")[, "fit"]
# add two more columns with predicted 95% CI uncertainty around the modeled DOC
conf_int <- predict(model, newdata = fDOM_DOC_join, interval = "confidence")
fDOM_DOC_join$lwr <- conf_int[, "lwr"]
fDOM_DOC_join$upr <- conf_int[, "upr"]
With the modeled data added to our joined data table, let's plot the resulting
modeled DOC w/ uncertainty on the same plotting field as the DOC measured from
water chemistry grab samples. We will make this plot using plotly so we can
zoom in to see how well the AOS-DOC and modeled continuous DOC match up.
# create plot
plotly::plot_ly(data=fDOM_DOC_join)%>%
# plot uncertainty as a ribbon
plotly::add_trace(x=~roundDate,y=~upr,name="95% CI",
type='scatter',mode='line',
line=list(color='lightgray'),legendgroup="95CI",
showlegend=F)%>%
plotly::add_trace(x=~roundDate,y=~lwr,name="95% CI",
type='scatter',mode='none',fill = 'tonexty',
fillcolor = 'lightgray',legendgroup="95CI")%>%
# plot modeled DOC timeseries
plotly::add_trace(x=~roundDate,y=~fit,name="Modeled DOC",
type='scatter',mode='line',
line=list(color='blue'))%>%
# plot grab sample DOC
plotly::add_trace(x=~roundDate,y=~analyteConcentration,name="Grab Sample DOC",
type='scatter',mode='markers',
marker=list(color='darkorange'))%>%
# format title, axes, and legend
plotly::layout(title="Dissolved Organic Carbon: Modelled & Grab Sample",
xaxis=list(title="Date"),
yaxis=list(title="DOC (mg/L)"),
legend=list(xanchor = 'center',
yanchor = 'top',
orientation = 'h',
x=0.5,y=-0.2))
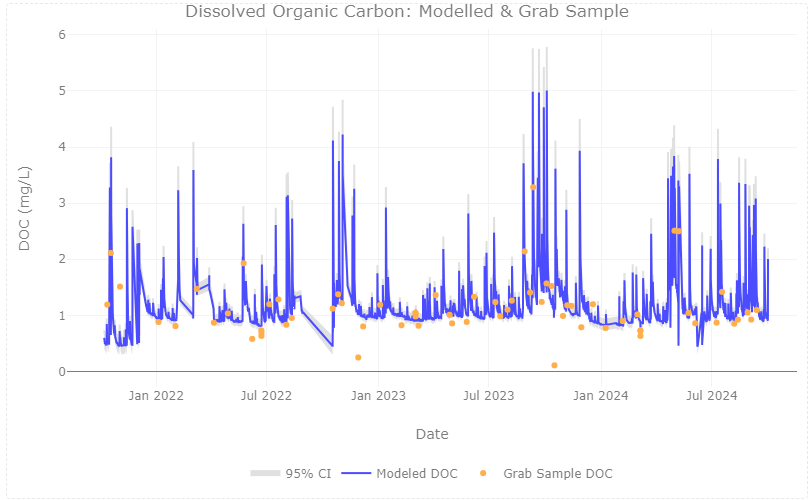
Discussion: This study shows the possibility of integrating AIS and AOS data to expand the temporal scale of estimated DOC in stream sites. At CUPE, estimated DOC matches well with grab sample DOC across mid-range values, with uncertainty increasing at the low and high ends of the timeseries. The relationship will continue to be expanded upon, following NEON’s flow-weighted sampling design.
The standardized nature of NEON data products and site designs allow for this and other analyses to be scaled across the observatory, revealing similarities and differences in the relationship across sites, and allowing for more detailed investigation of carbon dynamics in freshwater systems across the United States.
This analysis can be conducted for any of the 24 stream sites across the NEON
observatory simply by changing the site input variable in loadByProduct().
Try out this analysis at your favorite site! What environmental factors could
contribute to the quality of this relationship at different NEON stream sites?