Tutorial
Vector 01: Explore Shapefile Attributes & Plot Shapefile Objects by Attribute Value in R
Authors: Joseph Stachelek, Leah A. Wasser, Megan A. Jones
Last Updated: Apr 8, 2021
This tutorial explains what shapefile attributes are and how to work with shapefile attributes in R. It also covers how to identify and query shapefile attributes, as well as subset shapefiles by specific attribute values. Finally, we will review how to plot a shapefile according to a set of attribute values.
Learning Objectives
After completing this tutorial, you will be able to:
- Query shapefile attributes.
- Subset shapefiles using specific attribute values.
- Plot a shapefile, colored by unique attribute values.
Things You’ll Need To Complete This Tutorial
You will need the most current version of R and, preferably, RStudio loaded
on your computer to complete this tutorial.
Install R Packages
-
raster:
install.packages("raster") -
rgdal:
install.packages("rgdal") -
sp:
install.packages("sp")
More on Packages in R – Adapted from Software Carpentry.
Download Data
NEON Teaching Data Subset: Site Layout Shapefiles
These vector data provide information on the site characterization and infrastructure at the National Ecological Observatory Network's Harvard Forest field site. The Harvard Forest shapefiles are from the Harvard Forest GIS & Map archives. US Country and State Boundary layers are from the US Census Bureau.
Download DatasetSet Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded and unzipped data subsets.
An overview of setting the working directory in R can be found here.
R Script & Challenge Code: NEON data lessons often contain challenges that reinforce learned skills. If available, the code for challenge solutions is found in the downloadable R script of the entire lesson, available in the footer of each lesson page.
Shapefile Metadata & Attributes
When we import a shapefile into R, the readOGR() function automatically
stores metadata and attributes associated with the file.
Load the Data
To work with vector data in R, we can use the rgdal library. The raster
package also allows us to explore metadata using similar commands for both
raster and vector files.
We will import three shapefiles. The first is our AOI or area of
interest boundary polygon that we worked with in
Open and Plot Shapefiles in R.
The second is a shapefile containing the location of roads and trails within the
field site. The third is a file containing the Fisher tower location.
If you completed the Open and Plot Shapefiles in R. tutorial, you can skip this code.
# load packages
# rgdal: for vector work; sp package should always load with rgdal.
library(rgdal)
# raster: for metadata/attributes- vectors or rasters
library (raster)
# set working directory to data folder
# setwd("pathToDirHere")
# Import a polygon shapefile
aoiBoundary_HARV <- readOGR("NEON-DS-Site-Layout-Files/HARV",
"HarClip_UTMZ18", stringsAsFactors = T)
## OGR data source with driver: ESRI Shapefile
## Source: "/Users/olearyd/Git/data/NEON-DS-Site-Layout-Files/HARV", layer: "HarClip_UTMZ18"
## with 1 features
## It has 1 fields
## Integer64 fields read as strings: id
# Import a line shapefile
lines_HARV <- readOGR( "NEON-DS-Site-Layout-Files/HARV", "HARV_roads", stringsAsFactors = T)
## OGR data source with driver: ESRI Shapefile
## Source: "/Users/olearyd/Git/data/NEON-DS-Site-Layout-Files/HARV", layer: "HARV_roads"
## with 13 features
## It has 15 fields
# Import a point shapefile
point_HARV <- readOGR("NEON-DS-Site-Layout-Files/HARV",
"HARVtower_UTM18N", stringsAsFactors = T)
## OGR data source with driver: ESRI Shapefile
## Source: "/Users/olearyd/Git/data/NEON-DS-Site-Layout-Files/HARV", layer: "HARVtower_UTM18N"
## with 1 features
## It has 14 fields
Query Shapefile Metadata
Remember, as covered in Open and Plot Shapefiles in R., we can view metadata associated with an R object using:
-
class()- Describes the type of vector data stored in the object. -
length()- How many features are in this spatial object? - object
extent()- The spatial extent (geographic area covered by) features in the object. - coordinate reference system (
crs()) - The spatial projection that the data are in.
Let's explore the metadata for our point_HARV object.
# view class
class(x = point_HARV)
## [1] "SpatialPointsDataFrame"
## attr(,"package")
## [1] "sp"
# x= isn't actually needed; it just specifies which object
# view features count
length(point_HARV)
## [1] 1
# view crs - note - this only works with the raster package loaded
crs(point_HARV)
## CRS arguments:
## +proj=utm +zone=18 +datum=WGS84 +units=m +no_defs
# view extent- note - this only works with the raster package loaded
extent(point_HARV)
## class : Extent
## xmin : 732183.2
## xmax : 732183.2
## ymin : 4713265
## ymax : 4713265
# view metadata summary
point_HARV
## class : SpatialPointsDataFrame
## features : 1
## extent : 732183.2, 732183.2, 4713265, 4713265 (xmin, xmax, ymin, ymax)
## crs : +proj=utm +zone=18 +datum=WGS84 +units=m +no_defs
## variables : 14
## names : Un_ID, Domain, DomainName, SiteName, Type, Sub_Type, Lat, Long, Zone, Easting, Northing, Ownership, County, annotation
## value : A, 1, Northeast, Harvard Forest, Core, Advanced Tower, 42.5369, -72.17266, 18, 732183.193774, 4713265.041137, Harvard University, LTER, Worcester, C1
About Shapefile Attributes
Shapefiles often contain an associated database or spreadsheet of values called attributes that describe the vector features in the shapefile. You can think of this like a spreadsheet with rows and columns. Each column in the spreadsheet is an individual attribute that describes an object. Shapefile attributes include measurements that correspond to the geometry of the shapefile features.
For example, the HARV_Roads shapefile (lines_HARV object) contains an
attribute called TYPE. Each line in the shapefile has an associated TYPE
which describes the type of road (woods road, footpath, boardwalk, or
stone wall).
We can look at all of the associated data attributes by printing the contents of
the data slot with objectName@data. We can use the base R length
function to count the number of attributes associated with a spatial object too.
# just view the attributes & first 6 attribute values of the data
head(lines_HARV@data)
## OBJECTID_1 OBJECTID TYPE NOTES MISCNOTES RULEID
## 0 14 48 woods road Locust Opening Rd <NA> 5
## 1 40 91 footpath <NA> <NA> 6
## 2 41 106 footpath <NA> <NA> 6
## 3 211 279 stone wall <NA> <NA> 1
## 4 212 280 stone wall <NA> <NA> 1
## 5 213 281 stone wall <NA> <NA> 1
## MAPLABEL SHAPE_LENG LABEL BIKEHORSE RESVEHICLE
## 0 Locust Opening Rd 1297.35706 Locust Opening Rd Y R1
## 1 <NA> 146.29984 <NA> Y R1
## 2 <NA> 676.71804 <NA> Y R2
## 3 <NA> 231.78957 <NA> <NA> <NA>
## 4 <NA> 45.50864 <NA> <NA> <NA>
## 5 <NA> 198.39043 <NA> <NA> <NA>
## RECMAP Shape_Le_1 ResVehic_1
## 0 Y 1297.10617 R1 - All Research Vehicles Allowed
## 1 Y 146.29983 R1 - All Research Vehicles Allowed
## 2 Y 676.71807 R2 - 4WD/High Clearance Vehicles Only
## 3 <NA> 231.78962 <NA>
## 4 <NA> 45.50859 <NA>
## 5 <NA> 198.39041 <NA>
## BicyclesHo
## 0 Bicycles and Horses Allowed
## 1 Bicycles and Horses Allowed
## 2 Bicycles and Horses Allowed
## 3 <NA>
## 4 <NA>
## 5 <NA>
# how many attributes are in our vector data object?
length(lines_HARV@data)
## [1] 15
We can view the individual name of each attribute using the
names(lines_HARV@data) method in R. We could also view just the first 6 rows
of attribute values using head(lines_HARV@data).
Let's give it a try.
# view just the attribute names for the lines_HARV spatial object
names(lines_HARV@data)
## [1] "OBJECTID_1" "OBJECTID" "TYPE" "NOTES" "MISCNOTES"
## [6] "RULEID" "MAPLABEL" "SHAPE_LENG" "LABEL" "BIKEHORSE"
## [11] "RESVEHICLE" "RECMAP" "Shape_Le_1" "ResVehic_1" "BicyclesHo"
-
How many attributes do each have?
-
Who owns the site in the
point_HARVdata object? -
Which of the following is NOT an attribute of the
pointdata object?A) Latitude B) County C) Country
Explore Values within One Attribute
We can explore individual values stored within a particular attribute.
Again, comparing attributes to a spreadsheet or a data.frame is similar
to exploring values in a column. We can do this using the $ and the name of
the attribute: objectName$attributeName.
# view all attributes in the lines shapefile within the TYPE field
lines_HARV$TYPE
## [1] woods road footpath footpath stone wall stone wall stone wall
## [7] stone wall stone wall stone wall boardwalk woods road woods road
## [13] woods road
## Levels: boardwalk footpath stone wall woods road
# view unique values within the "TYPE" attributes
levels(lines_HARV@data$TYPE)
## [1] "boardwalk" "footpath" "stone wall" "woods road"
Notice that two of our TYPE attribute values consist of two separate words: stone wall and woods road. There are really four unique TYPE values, not six TYPE values.
Subset Shapefiles
We can use the objectName$attributeName syntax to select a subset of features
from a spatial object in R.
# select features that are of TYPE "footpath"
# could put this code into other function to only have that function work on
# "footpath" lines
lines_HARV[lines_HARV$TYPE == "footpath",]
## class : SpatialLinesDataFrame
## features : 2
## extent : 731954.5, 732232.3, 4713131, 4713726 (xmin, xmax, ymin, ymax)
## crs : +proj=utm +zone=18 +datum=WGS84 +units=m +no_defs
## variables : 15
## names : OBJECTID_1, OBJECTID, TYPE, NOTES, MISCNOTES, RULEID, MAPLABEL, SHAPE_LENG, LABEL, BIKEHORSE, RESVEHICLE, RECMAP, Shape_Le_1, ResVehic_1, BicyclesHo
## min values : 40, 91, footpath, NA, NA, 6, NA, 146.299844868, NA, Y, R1, Y, 146.299831389, R1 - All Research Vehicles Allowed, Bicycles and Horses Allowed
## max values : 41, 106, footpath, NA, NA, 6, NA, 676.71804248, NA, Y, R2, Y, 676.718065323, R2 - 4WD/High Clearance Vehicles Only, Bicycles and Horses Allowed
# save an object with only footpath lines
footpath_HARV <- lines_HARV[lines_HARV$TYPE == "footpath",]
footpath_HARV
## class : SpatialLinesDataFrame
## features : 2
## extent : 731954.5, 732232.3, 4713131, 4713726 (xmin, xmax, ymin, ymax)
## crs : +proj=utm +zone=18 +datum=WGS84 +units=m +no_defs
## variables : 15
## names : OBJECTID_1, OBJECTID, TYPE, NOTES, MISCNOTES, RULEID, MAPLABEL, SHAPE_LENG, LABEL, BIKEHORSE, RESVEHICLE, RECMAP, Shape_Le_1, ResVehic_1, BicyclesHo
## min values : 40, 91, footpath, NA, NA, 6, NA, 146.299844868, NA, Y, R1, Y, 146.299831389, R1 - All Research Vehicles Allowed, Bicycles and Horses Allowed
## max values : 41, 106, footpath, NA, NA, 6, NA, 676.71804248, NA, Y, R2, Y, 676.718065323, R2 - 4WD/High Clearance Vehicles Only, Bicycles and Horses Allowed
# how many features are in our new object
length(footpath_HARV)
## [1] 2
Our subsetting operation reduces the features count from 13 to 2. This means
that only two feature lines in our spatial object have the attribute
"TYPE=footpath".
We can plot our subsetted shapefiles.
# plot just footpaths
plot(footpath_HARV,
lwd=6,
main="NEON Harvard Forest Field Site\n Footpaths")
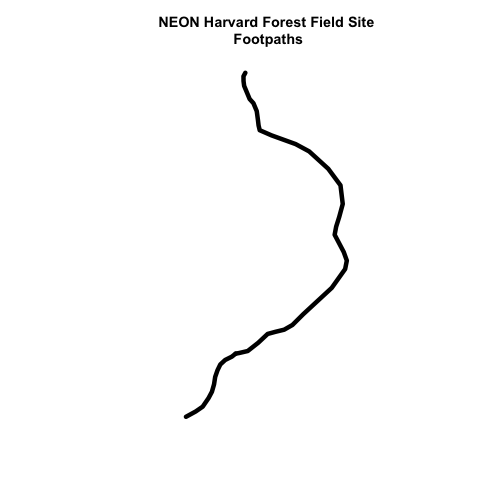
Interesting! Above, it appeared as if we had 2 features in our footpaths subset. Why does the plot look like there is only one feature?
Let's adjust the colors used in our plot. If we have 2 features in our vector
object, we can plot each using a unique color by assigning unique colors (col=)
to our features. We use the syntax
col="c("colorOne","colorTwo")
to do this.
# plot just footpaths
plot(footpath_HARV,
col=c("green","blue"), # set color for each feature
lwd=6,
main="NEON Harvard Forest Field Site\n Footpaths \n Feature one = blue, Feature two= green")
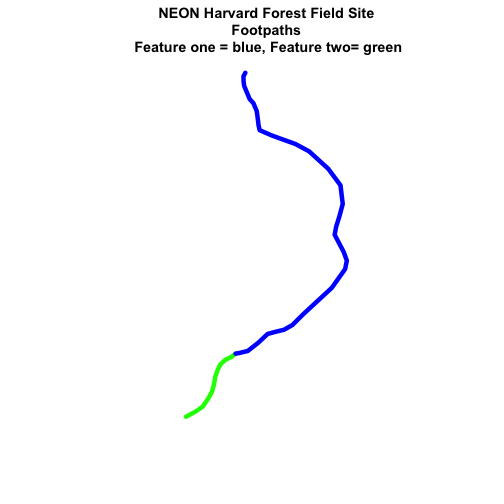
Now, we see that there are in fact two features in our plot!
-
boardwalkfrom the lines layer and plot it. -
stone wallfeatures from the lines layer and plot it.
For each plot, color each feature using a unique color.

Plot Lines by Attribute Value
To plot vector data with the color determined by a set of attribute values, the
attribute values must be class = factor. A factor is similar to a category.
- you can group vector objects by a particular category value - for example you
can group all lines of
TYPE=footpath. However, in R, a factor can also have a determined order.
By default, R will import spatial object attributes as factors.
# view the original class of the TYPE column
class(lines_HARV$TYPE)
## [1] "factor"
# view levels or categories - note that there are no categories yet in our data!
# the attributes are just read as a list of character elements.
levels(lines_HARV$TYPE)
## [1] "boardwalk" "footpath" "stone wall" "woods road"
# Convert the TYPE attribute into a factor
# Only do this IF the data do not import as a factor!
# lines_HARV$TYPE <- as.factor(lines_HARV$TYPE)
# class(lines_HARV$TYPE)
# levels(lines_HARV$TYPE)
# how many features are in each category or level?
summary(lines_HARV$TYPE)
## boardwalk footpath stone wall woods road
## 1 2 6 4
When we use plot(), we can specify the colors to use for each attribute using
the col= element. To ensure that R renders each feature by it's associated
factor / attribute value, we need to create a vector or colors - one for each
feature, according to it's associated attribute value / factor value.
To create this vector we can use the following syntax:
c("colorOne", "colorTwo","colorThree")[object$factor]
Note in the above example we have:
- a vector of colors - one for each factor value (unique attribute value)
- the attribute itself (
[object$factor]) of classfactor
Let's give this a try.
# Check the class of the attribute - is it a factor?
class(lines_HARV$TYPE)
## [1] "factor"
# how many "levels" or unique values does the factor have?
# view factor values
levels(lines_HARV$TYPE)
## [1] "boardwalk" "footpath" "stone wall" "woods road"
# count the number of unique values or levels
length(levels(lines_HARV$TYPE))
## [1] 4
# create a color palette of 4 colors - one for each factor level
roadPalette <- c("blue","green","grey","purple")
roadPalette
## [1] "blue" "green" "grey" "purple"
# create a vector of colors - one for each feature in our vector object
# according to its attribute value
roadColors <- c("blue","green","grey","purple")[lines_HARV$TYPE]
roadColors
## [1] "purple" "green" "green" "grey" "grey" "grey" "grey"
## [8] "grey" "grey" "blue" "purple" "purple" "purple"
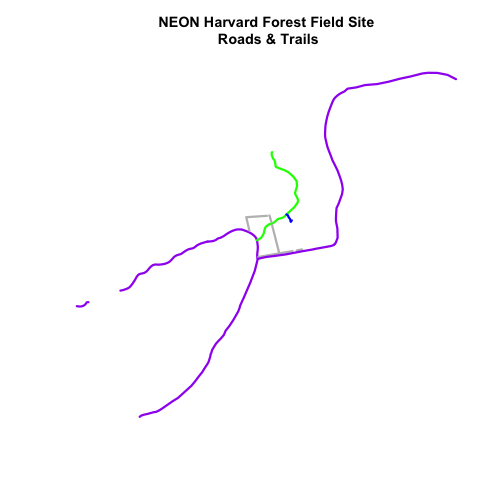
# plot the lines data, apply a diff color to each factor level)
plot(lines_HARV,
col=roadColors,
lwd=3,
main="NEON Harvard Forest Field Site\n Roads & Trails")
Adjust Line Width
We can also adjust the width of our plot lines using lwd. We can set all lines
to be thicker or thinner using lwd=.
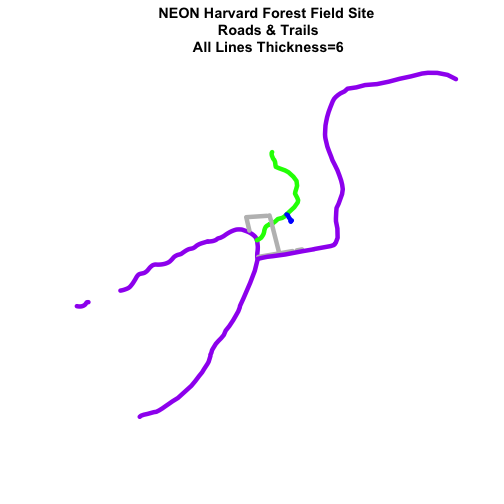
# make all lines thicker
plot(lines_HARV,
col=roadColors,
main="NEON Harvard Forest Field Site\n Roads & Trails\n All Lines Thickness=6",
lwd=6)
Adjust Line Width by Attribute
If we want a unique line width for each factor level or attribute category in our spatial object, we can use the same syntax that we used for colors, above:
lwd=c("widthOne", "widthTwo","widthThree")[object$factor]
Note that this requires the attribute to be of class factor. Let's give it a
try.
class(lines_HARV$TYPE)
## [1] "factor"
levels(lines_HARV$TYPE)
## [1] "boardwalk" "footpath" "stone wall" "woods road"
# create vector of line widths
lineWidths <- (c(1,2,3,4))[lines_HARV$TYPE]
# adjust line width by level
# in this case, boardwalk (the first level) is the widest.
plot(lines_HARV,
col=roadColors,
main="NEON Harvard Forest Field Site\n Roads & Trails \n Line width varies by TYPE Attribute Value",
lwd=lineWidths)
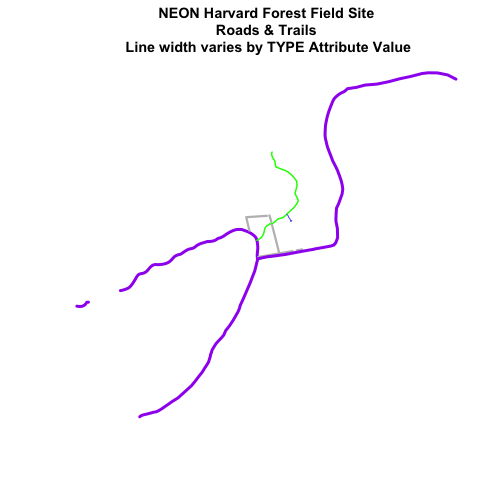
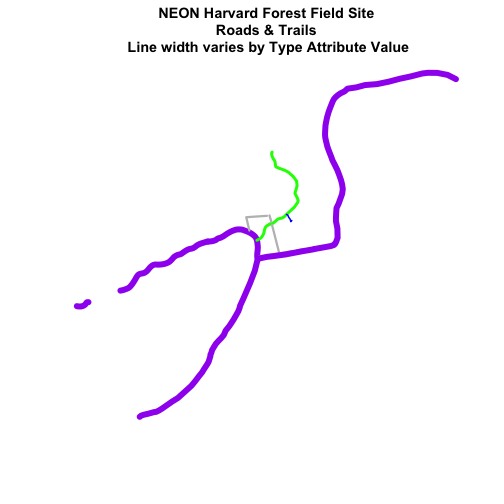
Create a plot of roads using the following line thicknesses:
- woods road lwd=8
- Boardwalks lwd = 2
- footpath lwd=4
- stone wall lwd=3
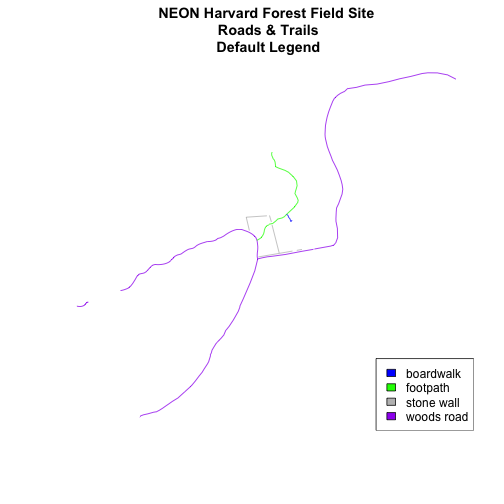
Add Plot Legend
We can add a legend to our plot too. When we add a legend, we use the following elements to specify labels and colors:
-
bottomright: We specify the location of our legend by using a default keyword. We could also usetop,topright, etc. -
levels(objectName$attributeName): Label the legend elements using the categories oflevelsin an attribute (e.g., levels(lines_HARV$TYPE) means use the levels boardwalk, footpath, etc). -
fill=: apply unique colors to the boxes in our legend.palette()is the default set of colors that R applies to all plots.
Let's add a legend to our plot.
plot(lines_HARV,
col=roadColors,
main="NEON Harvard Forest Field Site\n Roads & Trails\n Default Legend")
# we can use the color object that we created above to color the legend objects
roadPalette
## [1] "blue" "green" "grey" "purple"
# add a legend to our map
legend("bottomright", # location of legend
legend=levels(lines_HARV$TYPE), # categories or elements to render in
# the legend
fill=roadPalette) # color palette to use to fill objects in legend.
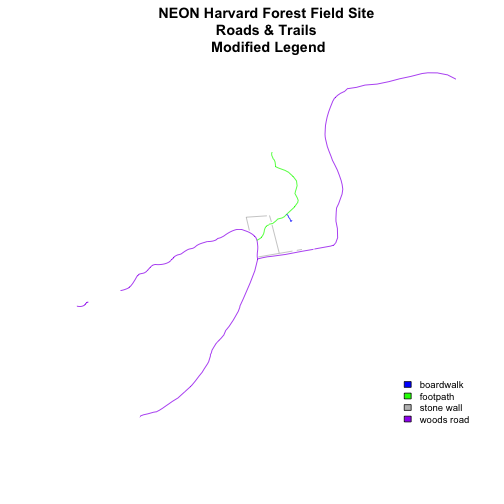
We can tweak the appearance of our legend too.
-
bty=n: turn off the legend BORDER -
cex: change the font size
Let's try it out.
plot(lines_HARV,
col=roadColors,
main="NEON Harvard Forest Field Site\n Roads & Trails \n Modified Legend")
# add a legend to our map
legend("bottomright",
legend=levels(lines_HARV$TYPE),
fill=roadPalette,
bty="n", # turn off the legend border
cex=.8) # decrease the font / legend size
We can modify the colors used to plot our lines by creating a new color vector directly in the plot code rather than creating a separate object.
col=(newColors)[lines_HARV$TYPE]
Let's try it!
# manually set the colors for the plot!
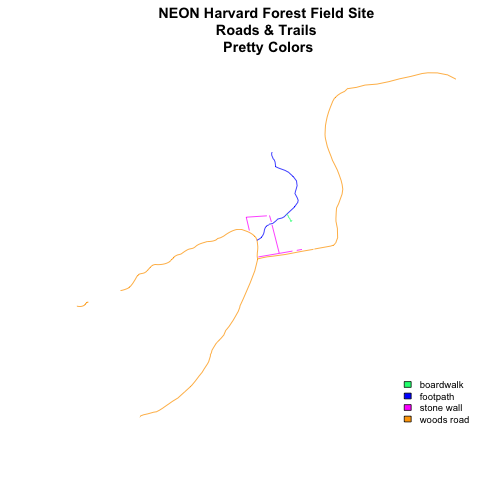
newColors <- c("springgreen", "blue", "magenta", "orange")
newColors
## [1] "springgreen" "blue" "magenta" "orange"
# plot using new colors
plot(lines_HARV,
col=(newColors)[lines_HARV$TYPE],
main="NEON Harvard Forest Field Site\n Roads & Trails \n Pretty Colors")
# add a legend to our map
legend("bottomright",
levels(lines_HARV$TYPE),
fill=newColors,
bty="n", cex=.8)
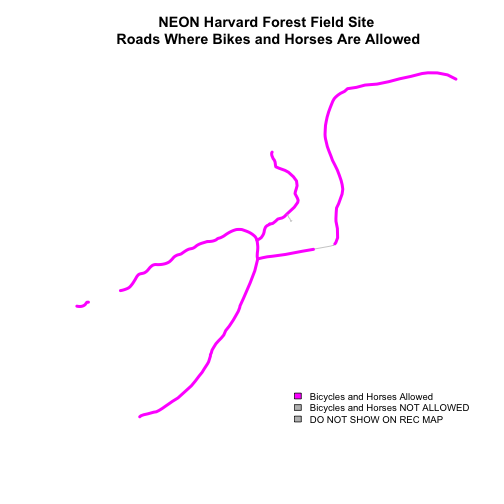
Be sure to add a title and legend to your map! You might consider a color palette that has all bike/horse-friendly roads displayed in a bright color. All other lines can be grey.
-
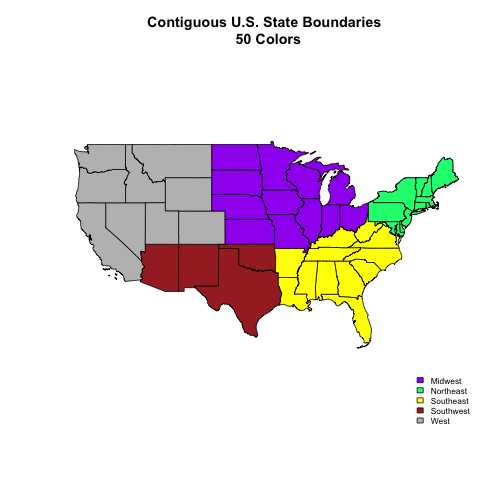
Create a map of the State boundaries in the United States using the data located in your downloaded data folder:
NEON-DS-Site-Layout-Files/US-Boundary-Layers\US-State-Boundaries-Census-2014. Apply a fill color to each state using itsregionvalue. Add a legend. -
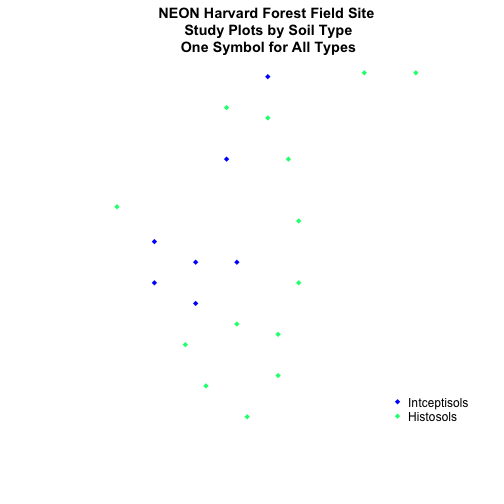
Using the
NEON-DS-Site-Layout-Files/HARV/PlotLocations_HARV.shpshapefile, create a map of study plot locations, with each point colored by the soil type (soilTypeOr). Question: How many different soil types are there at this particular field site? -
BONUS -- modify the field site plot above. Plot each point, using a different symbol. HINT: you can assign the symbol using
pch=value. You can create a vector object of symbols by factor level using the syntax syntax that we used above to create a vector of lines widths and colors:pch=c(15,17)[lines_HARV$soilTypeOr]. Type?pchto learn more about pch or use google to find a list of pch symbols that you can use in R.