Tutorial
Vector 02: Plot Multiple Shapefiles and Create Custom Legends in Base Plot in R
Authors: Joseph Stachelek, Leah A. Wasser, Megan A. Jones
Last Updated: Apr 8, 2021
This tutorial builds upon the previous tutorial, to work with shapefile attributes in R and explores how to plot multiple shapefiles using base R graphics. It then covers how to create a custom legend with colors and symbols that match your plot.
Learning Objectives
After completing this tutorial, you will be able to:
- Plot multiple shapefiles using base R graphics.
- Apply custom symbology to spatial objects in a plot in R.
- Customize a baseplot legend in R.
Things You’ll Need To Complete This Tutorial
You will need the most current version of R and preferably RStudio loaded
on your computer to complete this tutorial.
Install R Packages
-
raster:
install.packages("raster") -
rgdal:
install.packages("rgdal") -
sp:
install.packages("sp")
More on Packages in R – Adapted from Software Carpentry.
Download Data
NEON Teaching Data Subset: Site Layout Shapefiles
These vector data provide information on the site characterization and infrastructure at the National Ecological Observatory Network's Harvard Forest field site. The Harvard Forest shapefiles are from the Harvard Forest GIS & Map archives. US Country and State Boundary layers are from the US Census Bureau.
Download DatasetSet Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded and unzipped data subsets.
An overview of setting the working directory in R can be found here.
R Script & Challenge Code: NEON data lessons often contain challenges that reinforce learned skills. If available, the code for challenge solutions is found in the downloadable R script of the entire lesson, available in the footer of each lesson page.
Load the Data
To work with vector data in R, we can use the rgdal library. The raster
package also allows us to explore metadata using similar commands for both
raster and vector files.
We will import three shapefiles. The first is our AOI or area of
interest boundary polygon that we worked with in
Open and Plot Shapefiles in R.
The second is a shapefile containing the location of roads and trails within the
field site. The third is a file containing the Harvard Forest Fisher tower
location. These latter two we worked with in the
Explore Shapefile Attributes & Plot Shapefile Objects by Attribute Value in R tutorial.
# load packages
# rgdal: for vector work; sp package should always load with rgdal.
library(rgdal)
# raster: for metadata/attributes- vectors or rasters
library(raster)
# set working directory to data folder
# setwd("pathToDirHere")
# Import a polygon shapefile
aoiBoundary_HARV <- readOGR("NEON-DS-Site-Layout-Files/HARV",
"HarClip_UTMZ18", stringsAsFactors = T)
## OGR data source with driver: ESRI Shapefile
## Source: "/Users/olearyd/Git/data/NEON-DS-Site-Layout-Files/HARV", layer: "HarClip_UTMZ18"
## with 1 features
## It has 1 fields
## Integer64 fields read as strings: id
# Import a line shapefile
lines_HARV <- readOGR( "NEON-DS-Site-Layout-Files/HARV", "HARV_roads", stringsAsFactors = T)
## OGR data source with driver: ESRI Shapefile
## Source: "/Users/olearyd/Git/data/NEON-DS-Site-Layout-Files/HARV", layer: "HARV_roads"
## with 13 features
## It has 15 fields
# Import a point shapefile
point_HARV <- readOGR("NEON-DS-Site-Layout-Files/HARV",
"HARVtower_UTM18N", stringsAsFactors = T)
## OGR data source with driver: ESRI Shapefile
## Source: "/Users/olearyd/Git/data/NEON-DS-Site-Layout-Files/HARV", layer: "HARVtower_UTM18N"
## with 1 features
## It has 14 fields
Plot Data
In the Explore Shapefile Attributes & Plot Shapefile Objects by Attribute Value in R tutorial we created a plot where we customized the width of each line in a spatial object according to a factor level or category. To do this, we create a vector of colors containing a color value for EACH feature in our spatial object grouped by factor level or category.
# view the factor levels
levels(lines_HARV$TYPE)
## [1] "boardwalk" "footpath" "stone wall" "woods road"
# create vector of line width values
lineWidth <- c(2,4,3,8)[lines_HARV$TYPE]
# view vector
lineWidth
## [1] 8 4 4 3 3 3 3 3 3 2 8 8 8
# create a color palette of 4 colors - one for each factor level
roadPalette <- c("blue","green","grey","purple")
roadPalette
## [1] "blue" "green" "grey" "purple"
# create a vector of colors - one for each feature in our vector object
# according to its attribute value
roadColors <- c("blue","green","grey","purple")[lines_HARV$TYPE]
roadColors
## [1] "purple" "green" "green" "grey" "grey" "grey" "grey"
## [8] "grey" "grey" "blue" "purple" "purple" "purple"
# create vector of line width values
lineWidth <- c(2,4,3,8)[lines_HARV$TYPE]
# view vector
lineWidth
## [1] 8 4 4 3 3 3 3 3 3 2 8 8 8
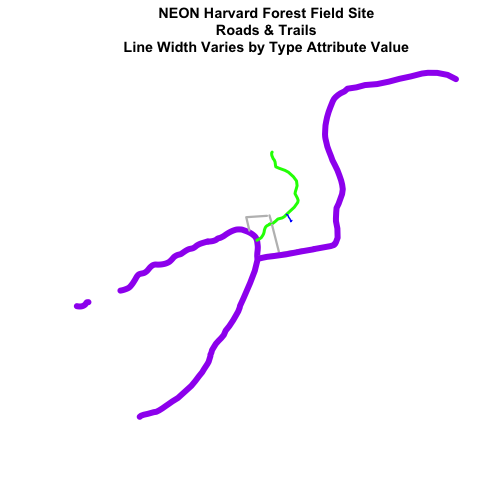
# in this case, boardwalk (the first level) is the widest.
plot(lines_HARV,
col=roadColors,
main="NEON Harvard Forest Field Site\n Roads & Trails \nLine Width Varies by Type Attribute Value",
lwd=lineWidth)
Add Plot Legend
In the the previous tutorial, we also learned how to add a basic legend to our plot.
-
bottomright: We specify the location of our legend by using a default keyword. We could also usetop,topright, etc. -
levels(objectName$attributeName): Label the legend elements using the categories oflevelsin an attribute (e.g., levels(lines_HARV$TYPE) means use the levels boardwalk, footpath, etc). -
fill=: apply unique colors to the boxes in our legend.palette()is the default set of colors that R applies to all plots.
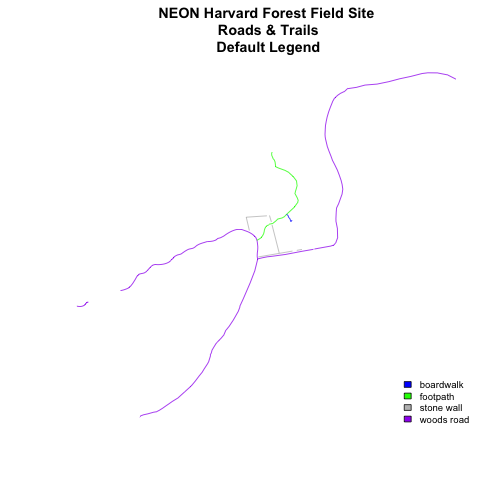
Let's add a legend to our plot.
plot(lines_HARV,
col=roadColors,
main="NEON Harvard Forest Field Site\n Roads & Trails\n Default Legend")
# we can use the color object that we created above to color the legend objects
roadPalette
## [1] "blue" "green" "grey" "purple"
# add a legend to our map
legend("bottomright",
legend=levels(lines_HARV$TYPE),
fill=roadPalette,
bty="n", # turn off the legend border
cex=.8) # decrease the font / legend size
However, what if we want to create a more complex plot with many shapefiles and unique symbols that need to be represented clearly in a legend?
Plot Multiple Vector Layers
Now, let's create a plot that combines our tower location (point_HARV),
site boundary (aoiBoundary_HARV) and roads (lines_HARV) spatial objects. We
will need to build a custom legend as well.
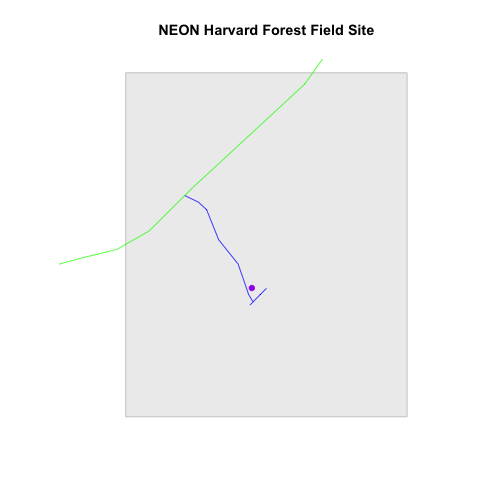
To begin, create a plot with the site boundary as the first layer. Then layer
the tower location and road data on top using add=TRUE.
# Plot multiple shapefiles
plot(aoiBoundary_HARV,
col = "grey93",
border="grey",
main="NEON Harvard Forest Field Site")
plot(lines_HARV,
col=roadColors,
add = TRUE)
plot(point_HARV,
add = TRUE,
pch = 19,
col = "purple")
# assign plot to an object for easy modification!
plot_HARV<- recordPlot()
Customize Your Legend
Next, let's build a custom legend using the symbology (the colors and symbols) that we used to create the plot above. To do this, we will need to build three things:
- A list of all "labels" (the text used to describe each element in the legend to use in the legend.
- A list of colors used to color each feature in our plot.
- A list of symbols to use in the plot. NOTE: we have a combination of points, lines and polygons in our plot. So we will need to customize our symbols!
Let's create objects for the labels, colors and symbols so we can easily reuse them. We will start with the labels.
# create a list of all labels
labels <- c("Tower", "AOI", levels(lines_HARV$TYPE))
labels
## [1] "Tower" "AOI" "boardwalk" "footpath" "stone wall"
## [6] "woods road"
# render plot
plot_HARV
# add a legend to our map
legend("bottomright",
legend=labels,
bty="n", # turn off the legend border
cex=.8) # decrease the font / legend size
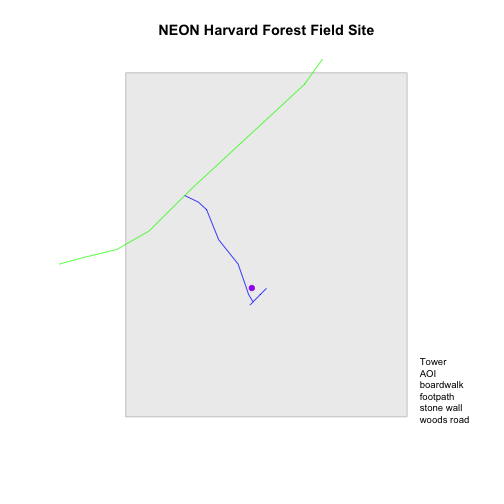
Now we have a legend with the labels identified. Let's add colors to each legend element next. We can use the vectors of colors that we created earlier to do this.
# we have a list of colors that we used above - we can use it in the legend
roadPalette
## [1] "blue" "green" "grey" "purple"
# create a list of colors to use
plotColors <- c("purple", "grey", roadPalette)
plotColors
## [1] "purple" "grey" "blue" "green" "grey" "purple"
# render plot
plot_HARV
# add a legend to our map
legend("bottomright",
legend=labels,
fill=plotColors,
bty="n", # turn off the legend border
cex=.8) # decrease the font / legend size
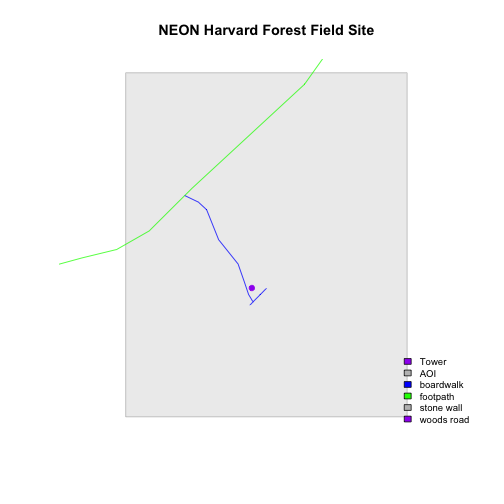
Great, now we have a legend! However, this legend uses boxes to symbolize each
element in the plot. It might be better if the lines were symbolized as a line
and the points were symbolized as a point symbol. We can customize this using
pch= in our legend: 16 is a point symbol, 15 is a box.
# create a list of pch values
# these are the symbols that will be used for each legend value
# ?pch will provide more information on values
plotSym <- c(16,15,15,15,15,15)
plotSym
## [1] 16 15 15 15 15 15
# Plot multiple shapefiles
plot_HARV
# to create a custom legend, we need to fake it
legend("bottomright",
legend=labels,
pch=plotSym,
bty="n",
col=plotColors,
cex=.8)
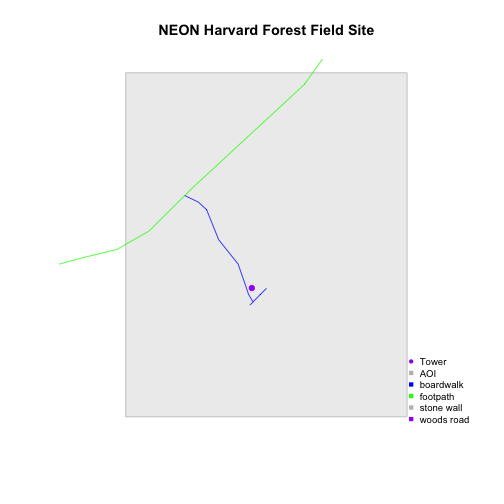
Now we've added a point symbol to represent our point element in the plot. However
it might be more useful to use line symbols in our legend
rather than squares to represent the line data. We can create line symbols,
using lty = (). We have a total of 6 elements in our legend:
- A Tower Location
- An Area of Interest (AOI)
- and 4 Road types (levels)
The lty list designates, in order, which of those elements should be
designated as a line (1) and which should be designated as a symbol (NA).
Our object will thus look like lty = c(NA,NA,1,1,1,1). This tells R to only use a
line element for the 3-6 elements in our legend.
Once we do this, we still need to modify our pch element. Each line element
(3-6) should be represented by a NA value - this tells R to not use a
symbol, but to instead use a line.
# create line object
lineLegend = c(NA,NA,1,1,1,1)
lineLegend
## [1] NA NA 1 1 1 1
plotSym <- c(16,15,NA,NA,NA,NA)
plotSym
## [1] 16 15 NA NA NA NA
# plot multiple shapefiles
plot_HARV
# build a custom legend
legend("bottomright",
legend=labels,
lty = lineLegend,
pch=plotSym,
bty="n",
col=plotColors,
cex=.8)
![Roads and tower location at NEON Harvard Forest Field Site with color and a modified legend varied by attribute type; each symbol on the legend corresponds to the shapefile type [i.e., tower = point, roads = lines].](https://raw.githubusercontent.com/NEONScience/NEON-Data-Skills/main/tutorials/R/Geospatial-skills/intro-vector-r/02-plot-multiple-shapefiles-custom-legend/rfigs/refine-legend-1.png)
-
Using the
NEON-DS-Site-Layout-Files/HARV/PlotLocations_HARV.shpshapefile, create a map of study plot locations, with each point colored by the soil type (soilTypeOr). How many different soil types are there at this particular field site? Overlay this layer on top of thelines_HARVlayer (the roads). Create a custom legend that applies line symbols to lines and point symbols to the points. -
Modify the plot above. Tell R to plot each point, using a different symbol of
pchvalue. HINT: to do this, create a vector object of symbols by factor level using the syntax described above for line width:c(15,17)[lines_HARV$soilTypeOr]. Overlay this on top of the AOI Boundary. Create a custom legend.
![Roads and study plots at NEON Harvard Forest Field Site with color and a modified legend varied by attribute type; each symbol on the legend corresponds to the shapefile type [i.e., soil plots = points, roads = lines].](https://raw.githubusercontent.com/NEONScience/NEON-Data-Skills/main/tutorials/R/Geospatial-skills/intro-vector-r/02-plot-multiple-shapefiles-custom-legend/rfigs/challenge-code-plot-color-1.png)
![Roads and study plots at NEON Harvard Forest Field Site with color and a modified legend varied by attribute type; each symbol on the legend corresponds to the shapefile type [i.e., soil plots = points, roads = lines], and study plots symbols vary by soil type.](https://raw.githubusercontent.com/NEONScience/NEON-Data-Skills/main/tutorials/R/Geospatial-skills/intro-vector-r/02-plot-multiple-shapefiles-custom-legend/rfigs/challenge-code-plot-color-2.png)