Tutorial
Hierarchical Data Formats - What is HDF5?
Authors: Leah A. Wasser
Last Updated: Apr 10, 2025
Learning Objectives
After completing this tutorial, you will be able to:
- Explain what the Hierarchical Data Format (HDF5) is.
- Describe the key benefits of the HDF5 format, particularly related to big data.
- Describe both the types of data that can be stored in HDF5 and how it can be stored/structured.
About Hierarchical Data Formats - HDF5
The Hierarchical Data Format version 5 (HDF5), is an open source file format that supports large, complex, heterogeneous data. HDF5 uses a "file directory" like structure that allows you to organize data within the file in many different structured ways, as you might do with files on your computer. The HDF5 format also allows for embedding of metadata making it self-describing.
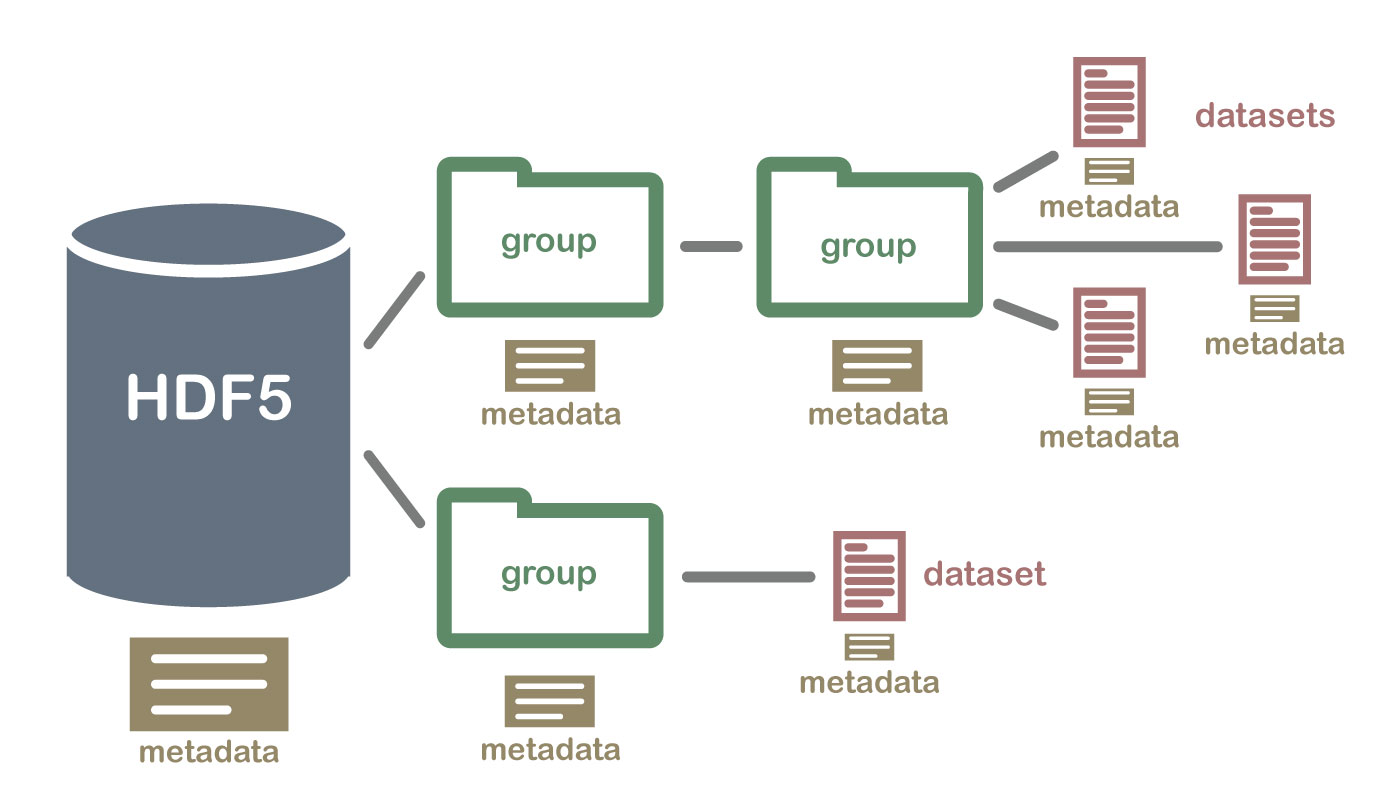
Hierarchical Structure - A file directory within a file
The HDF5 format can be thought of as a file system contained and described
within one single file. Think about the files and folders stored on your computer.
You might have a data directory with some temperature data for multiple field
sites. These temperature data are collected every minute and summarized on an
hourly, daily and weekly basis. Within one HDF5 file, you can store a similar
set of data organized in the same way that you might organize files and folders
on your computer. However in a HDF5 file, what we call "directories" or "folders"
on our computers, are called groups and what we call files on our
computer are called datasets.
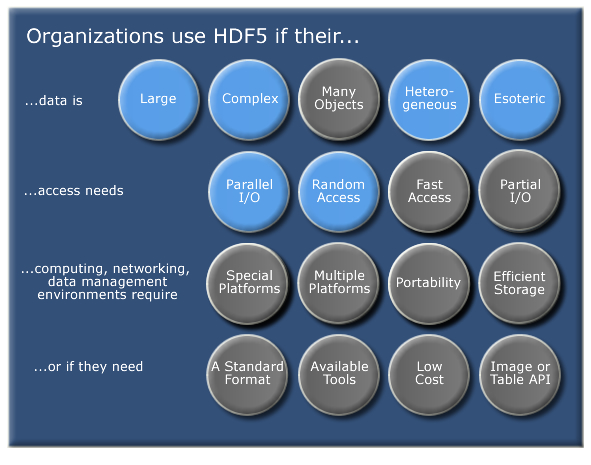
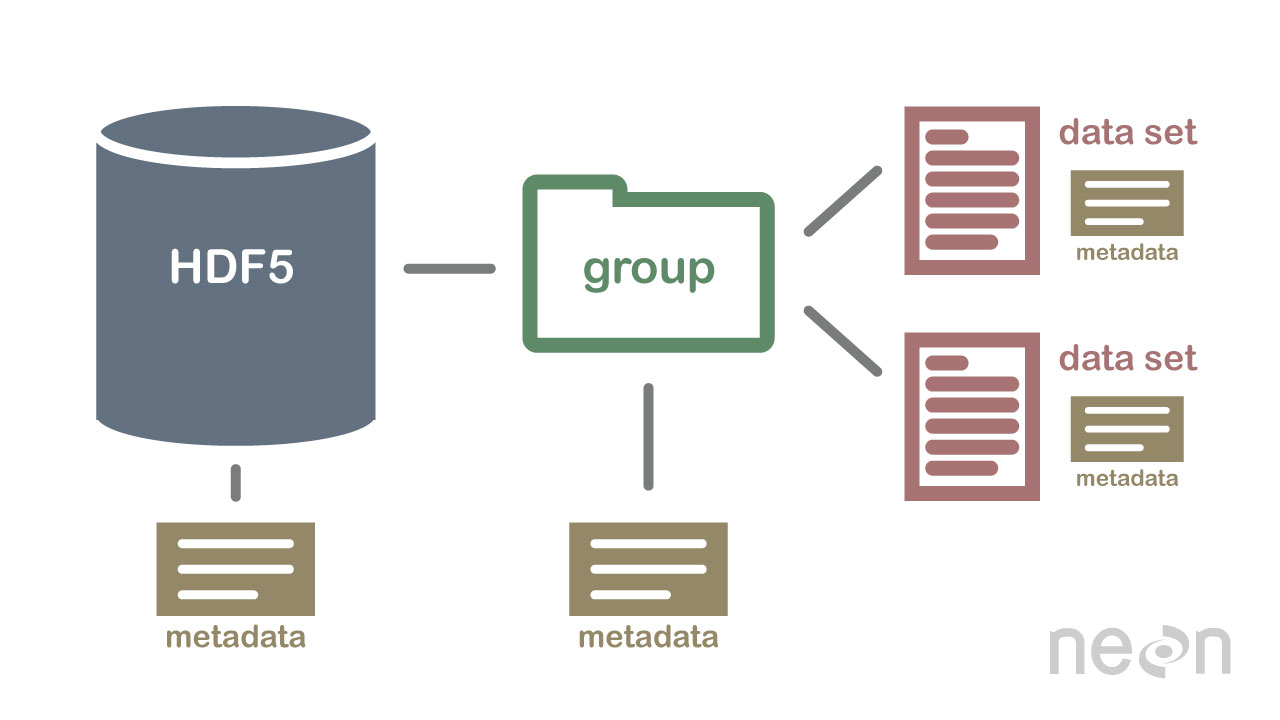
2 Important HDF5 Terms
- Group: A folder like element within an HDF5 file that might contain other groups OR datasets within it.
- Dataset: The actual data contained within the HDF5 file. Datasets are often (but don't have to be) stored within groups in the file.
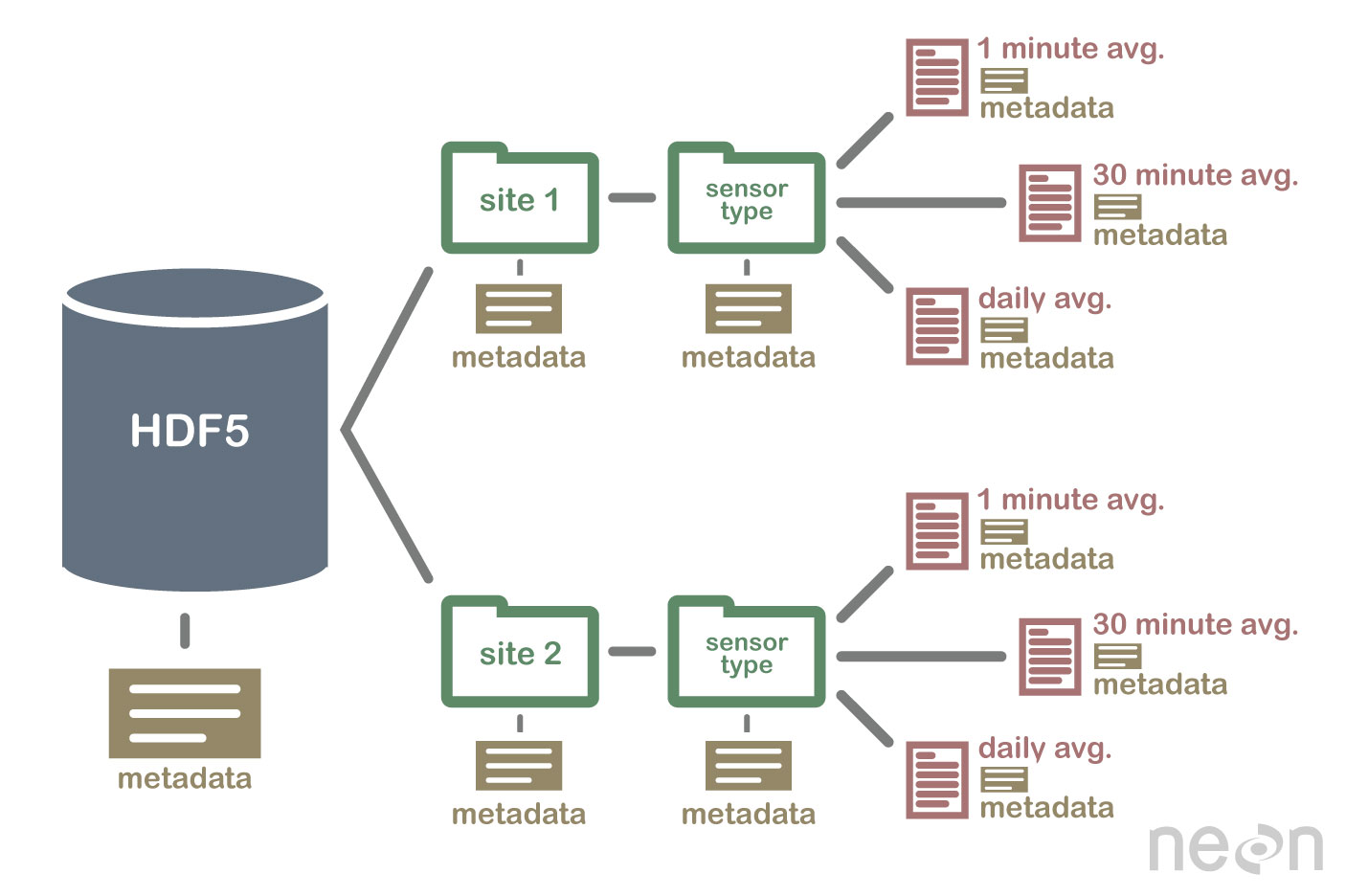
An HDF5 file containing datasets, might be structured like this:
HDF5 is a Self Describing Format
HDF5 format is self describing. This means that each file, group and dataset can have associated metadata that describes exactly what the data are. Following the example above, we can embed information about each site to the file, such as:
- The full name and X,Y location of the site
- Description of the site.
- Any documentation of interest.
Similarly, we might add information about how the data in the dataset were collected, such as descriptions of the sensor used to collect the temperature data. We can also attach information, to each dataset within the site group, about how the averaging was performed and over what time period data are available.
One key benefit of having metadata that are attached to each file, group and
dataset, is that this facilitates automation without the need for a separate
(and additional) metadata document. Using a programming language, like R or
Python, we can grab information from the metadata that are already associated
with the dataset, and which we might need to process the dataset.
Compressed & Efficient subsetting
The HDF5 format is a compressed format. The size of all data contained within
HDF5 is optimized which makes the overall file size smaller. Even when
compressed, however, HDF5 files often contain big data and can thus still be
quite large. A powerful attribute of HDF5 is data slicing, by which a
particular subsets of a dataset can be extracted for processing. This means that
the entire dataset doesn't have to be read into memory (RAM); very helpful in
allowing us to more efficiently work with very large (gigabytes or more) datasets!
Heterogeneous Data Storage
HDF5 files can store many different types of data within in the same file. For example, one group may contain a set of datasets to contain integer (numeric) and text (string) data. Or, one dataset can contain heterogeneous data types (e.g., both text and numeric data in one dataset). This means that HDF5 can store any of the following (and more) in one file:
- Temperature, precipitation and PAR (photosynthetic active radiation) data for a site or for many sites
- A set of images that cover one or more areas (each image can have specific spatial information associated with it - all in the same file)
- A multi or hyperspectral spatial dataset that contains hundreds of bands.
- Field data for several sites characterizing insects, mammals, vegetation and meteorology.
- A set of images that cover one or more areas (each image can have unique spatial information associated with it)
- And much more!
Open Format
The HDF5 format is open and free to use. The supporting libraries (and a free
viewer), can be downloaded from the
HDF Group
website. As such, HDF5 is widely supported in a host of programs, including
open source programming languages like R and Python, and commercial
programming tools like Matlab and IDL. Spatial data that are stored in HDF5
format can be used in GIS and imaging programs including QGIS, ArcGIS, and
ENVI.
Summary Points - Benefits of HDF5
- Self-Describing The datasets with an HDF5 file are self describing. This allows us to efficiently extract metadata without needing an additional metadata document.
- Supporta Heterogeneous Data: Different types of datasets can be contained within one HDF5 file.
- Supports Large, Complex Data: HDF5 is a compressed format that is designed to support large, heterogeneous, and complex datasets.
- Supports Data Slicing: "Data slicing", or extracting portions of the dataset as needed for analysis, means large files don't need to be completely read into the computers memory or RAM.
-
Open Format - wide support in the many tools: Because the HDF5 format is
open, it is supported by a host of programming languages and tools, including
open source languages like R and
Pythonand open GIS tools likeQGIS.