Tutorial
Publish Code - From R Markdown to HTML with knitr
Last Updated: Apr 8, 2021
Authors:
Last Updated: Apr 8, 2021
In this tutorial, we will cover the R knitr package that is used to convert
R Markdown into a rendered document (HTML, PDF, etc).
At the end of this activity, you will:
You will need the most current version of R and, preferably, RStudio loaded on your computer to complete this tutorial.
install.packages("knitr")
install.packages("rmarkdown")
The knitr package allow us to:
To complete this tutorial you need:
knitr package to complete this tutorial. If you need help installing
packages, visit
the R packages tutorial.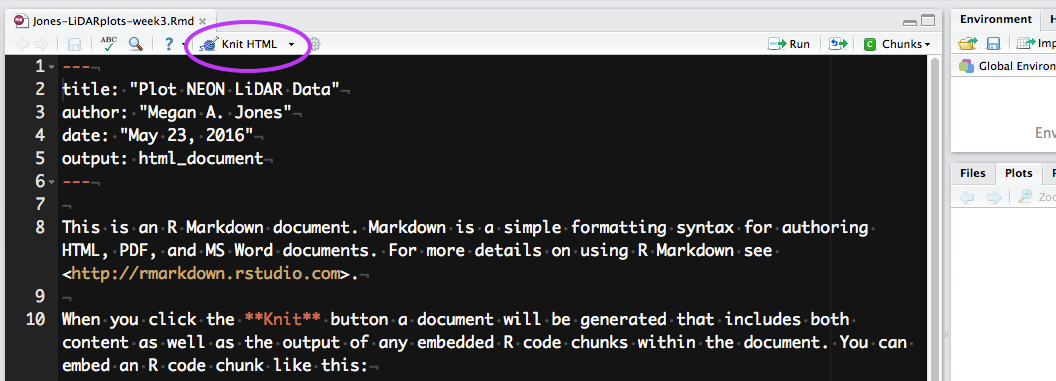
To knit in RStudio, click the knit pull down button. You want to use the
knit HTML for this lesson.
When you click the Knit HTML button, a window will open in your console titled R Markdown. This pane shows the knitting progress. The output (HTML in this case) file will automatically be saved in the current working directory. If there is an error in the code, an error message will appear with a line number in the R Console to help you diagnose the problem.
Knit the .Rmd file that you built in the last tutorial. What does it look like?
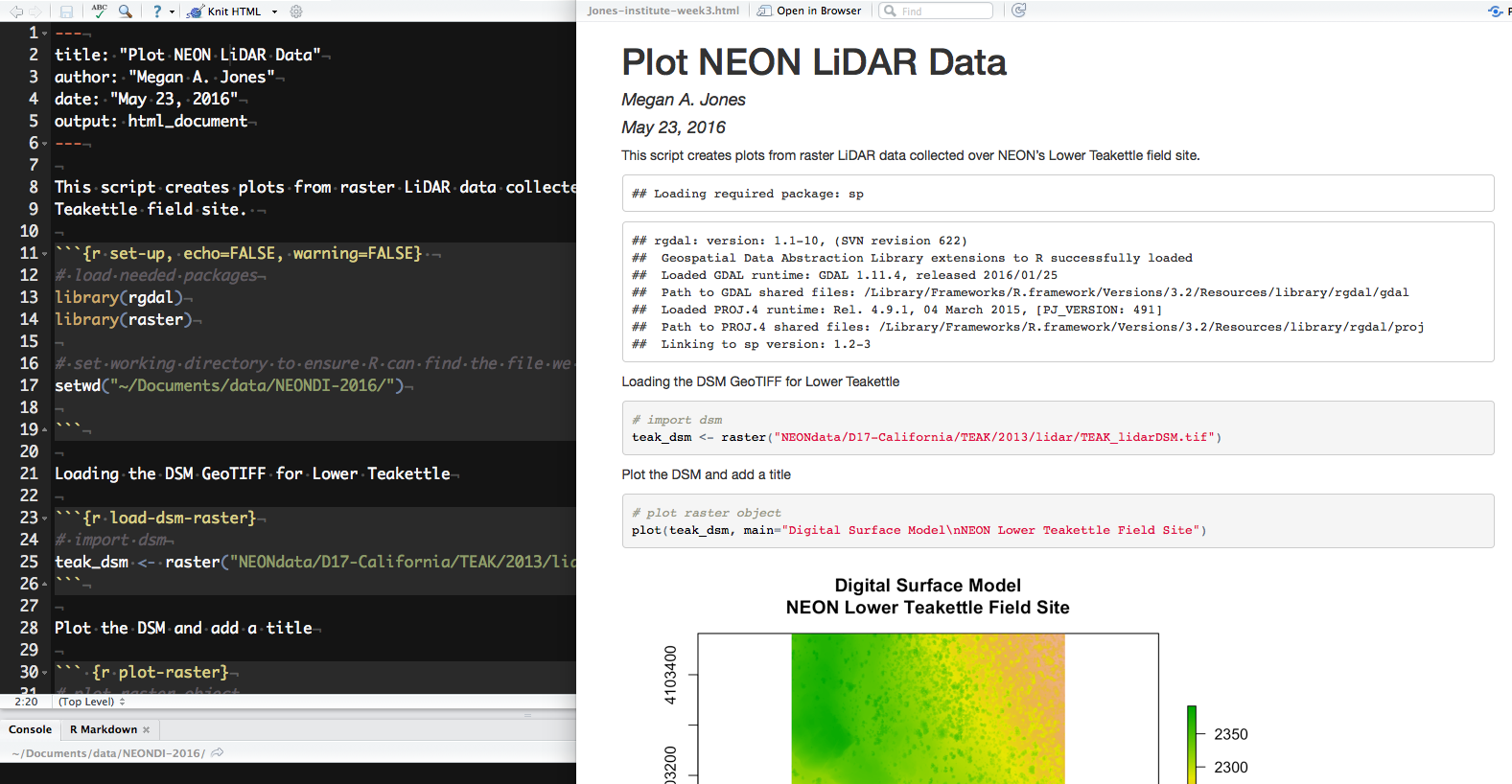
When knitting is complete, the new HTML file produced will automatically open.
Notice that information from the YAML header (title, author, date) is printed at the top of the HTML document. Then the HTML shows the text, code, and results of the code that you included in the RMD document.